
On Metrics for Measuring Fragmentation of Federation over SPARQL
Endpoints
Nur Aini Rakhmawati, Marcel Karnstedt, Michael Hausenblas and Stefan Decker
INSIGHT Centre, National University of Ireland, Galway, Ireland
Keywords:
Linked Data, Data Distribution, Federated SPARQL Query, SPARQL Endpoint.
Abstract:
Processing a federated query in Linked Data is challenging because it needs to consider the number of sources,
the source locations as well as heterogeneous system such as hardware, software and data structure and distri-
bution. In this work, we investigate the relationship between the data distribution and the communication cost
in a federated SPARQL query framework. We introduce the spreading factor as a dataset metric for computing
the distribution of classes and properties throughout a set of data sources. To observe the relationship between
the spreading factor and the communication cost, we generate 9 datasets by using several data fragmentation
and allocation strategies. Our experimental results showed that the spreading factor is correlated with the com-
munication cost between a federated engine and the SPARQL endpoints . In terms of partitioning strategies,
partitioning triples based on the properties and classes can minimize the communication cost. However, such
partitioning can also reduce the performance of SPARQL endpoint within the federation framework.
1 INTRODUCTION
Processing a federated query in the Linked Data is
challenging because it needs to consider the number
of the sources, the source locations and heterogeneous
system such as the hardware, the software and the data
structure and the distribution. A federated SPARQL
query can be easily formulated by using the SERVICE
keyword. Nevertheless, determining the datasource
address that follows SERVICE keywords can be an
obstacle in writing a query because prior knowledge
data is required. To address this issue, several ap-
proaches (Rakhmawati et al., 2013) have been devel-
oped with the objective of hiding SERVICE keyword
and data sources location from the user. In these ap-
proaches, the federated engines receive a query from
the user, parse the query into sub queries, decide
the location of each sub query and distribute the sub
queries to the relevant sources. A sub query can be
delivered to more than one data source if the desired
answer occurs in the multiple sources. Thus, the dis-
tribution of the data can affect the federation perfor-
mance (Rakhmawati and Hausenblas, 2012). As an
example, consider two datasets shown in Figure 1.
Each dataset contains a list of personal information
using the FOAF(http://xmlns.com/foaf/spec/) vocab-
ulary. If the user asks for the list of all person names,
the federated engine must send a query to all data-
Figure 1: Example of Federated SPARQL Query Involving
Many Datasets.
sources. Consequently, the communication cost be-
tween the federated engine and data sources would be
expensive.
In this study, we investigate the effect of data dis-
tribution on the federated engine performance. We
propose two composite metrics to calculate the pres-
ence of classes and properties across datasets. These
metrics can provide insight into the data distribu-
tion in the dataset which ultimately, it can determine
the communication cost between the federated en-
gine and SPARQL Endpoints. In order to evaluate
our metrics, we use several fragmentation and allo-
cation strategies to generate different shapes of data
distribution. After that, we run a static query set over
those data distributions. Our data distribution strate-
gies could be useful for benchmarking and controlled
systems such as organization system, but they can
not be address the problem in the federated Linked
99
Aini Rakhmawati N., Karnstedt M., Hausenblas M. and Decker S..
On Metrics for Measuring Fragmentation of Federation over SPARQL Endpoints.
DOI: 10.5220/0004858000990106
In Proceedings of the 10th International Conference on Web Information Systems and Technologies (WEBIST-2014), pages 99-106
ISBN: 978-989-758-024-6
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

Open Data environment because the Linked Data pub-
lisher has the power to control the dataset genera-
tion. The existing evaluations for assessing the feder-
ation over SPARQL endpoints (Montoya et al., 2012;
Schwarte et al., 2012) usually run their experiment
over different datasets and different query sets. In
fact, the performance of the federated engine is influ-
enced by both dataset and query set. As a result, the
performance results may vary. For benchmarking, a
better comparison of federated engines performance
can made with either static query sets over different
datasets or static dataset with various query sets.
We only perform our observation on federation
over SPARQL endpoints. Query with a SERVICE
keyword is also out of the scope of our study be-
cause the query only goes to the specified source. In
other words, the data distribution does not influence
the performance of the federation engine in that query.
Our contributions can be stated as follows: 1) We in-
vestigate the effects of data fragmentation and allo-
cation on the communication cost of the Federated
SPARQL query. 2) We introduce the spreading fac-
tor as a metric for calculating the distribution of data
across a dataset. In addition, we present the relation-
ship between the spreading factor and the communi-
cation cost of federated SPARQL queries. 3) Lastly,
we create datasets for evaluating the spreading factor
metric drawing from the real datasets. In particular,
we provide datasets and a dataset generator that can
be useful for benchmarking purpose.
2 RELATED WORKS
Primitive data metrics such as the number of triples,
the number of literals are not sufficiently represen-
tative to reveal the essential characteristics of the
datasets. Thus, Duan (Duan et al., 2011) introduced
a structuredness notion. Since this notion is applied
to a single RDF repository, it is not suitable for feder-
ated SPARQL queries which should consider the data
allocation in each repository as well as the number of
data sources involved in the dataset.
There are several data partitioning approaches for
RDF data clustering repository such as vertical parti-
tioning (Abadi et al., 2007) and Property Table par-
titioning (Huang et al., 2011). However, the commu-
nication in the RDF data clustering is totally differ-
ent than the communication in the federated SPARQL
query. In data clustering, several machines need
to communicate with each other in order to execute
a query, whereas in the federated SPARQL query,
there is no interaction amongst SPARQL endpoints.
The mediator has a role to communicate to each
SPARQL endpoint during query execution in the fed-
erated SPARQL query. Nevertheless, we apply RDF
data clustering strategies to generate the datasets for
evaluation.
The existing evaluations of the federation frame-
works used data partitioning in their experiment by
adopting data clustering strategies. Prasser (Prasser
et al., 2012) implemented three partitions: naturally-
partitioned, horizontally-partitioned and randomly-
partitioned. Fedbench(Schmidt et al., 2011) divided
the SP2B (Schmidt et al., 2009) dataset into sev-
eral partitions to run one of their evaluations. Our
prior work (Rakhmawati and Hausenblas, 2012) ob-
served the impact of data distribution on federated
query execution which particularly focus on the num-
ber of sources involved, the number of links and the
populated entities in several sources. In this work,
we extend our previous evaluation by implementing
more data partitioning schemes and we investigate
the effect of the distribution of classes and properties
throughout the dataset partitions on the performance
of federated SPARQL query.
3 SPREADING FACTOR OF
DATASET
Federated engines generally use a data catalogue to
predict the most relevant sources for a sub query.
The data catalogue mostly consists of a list of pred-
icates and classes. Apart from deciding the destina-
tion of the sub queries, a data catalogue can help fed-
erated engine generate set of query execution plans.
Hence, we consider computing the Spreading factor
of dataset to analyse the distribution of classes and
properties throughout the dataset. We initially define
the dataset used in this paper as follows:
Definition 1. Dataset D is a finite set of data sources
d. In the context of federation over SPARQL end-
points, d denotes a set of triple statements t that
can be accessed by a SPARQL endpoint. For each
SPARQL endpoint, there exists multiple RDF graphs.
In our work, we ignore the existence of graphs,
because we are only interested in the occurrences of
properties and classes in the SPARQL endpoint.
Definition 2. Let U be the set of all URIs, B be the set
of all BlankNodes, L be the set of all Literals, then a
triple t = (s, p,o) ∈ (U ∪ B) ×U × (U ∪ L ∪ B) where
s is the subject, p is the predicate and o is the object
of triple t.
Later on, we determine the property and the class
in the dataset as follows:
WEBIST2014-InternationalConferenceonWebInformationSystemsandTechnologies
100

Definition 3. Suppose d is a datasource in the dataset
D, then the set P
d
(d,D) of properties p in the source
d is defined as P
d
(d,D) = {p|∃(s, p,o) ∈ d ∧ d ∈ D}
and the set P(D) of properties p in the dataset D is
defined as P(D) = {p|p ∈ P
d
(d,D) ∧ d ∈ D}
Definition 4. Suppose d is a datasource in the dataset
D, then the set C
d
(d,D) of classes c in the source d
is defined as C
d
(d,D) = {c|∃(s,rd f type,c) ∈ d ∧ d ∈
D} and the set of classes c in the dataset D is defined
as C(D) = {c|c ∈ C
d
(d,D) ∧ d ∈ D}
Given two datasets D = {d
1
,d
2
} as shown in
Figure 1. Then P
d
(d
1
,D) = {rdf:type,foaf:name},
P
d
(d
2
,D) = P(D) = {rdf:type, foaf:name,foaf:mbox}
and C
d
(d
1
,D) = C
d
(d
2
,D) = C(D) = {foaf:person}.
3.1 Spreading Factor of Dataset
With the above definitions of class, property and
dataset, now we can describe how we calculate the
spreading factor. The spreading factor of the dataset
is based on whether or not classes and properties oc-
cur. Note that, we do not count the number of times
a class and property that are found in the source d be-
cause the federated engine usually relies on the pres-
ence of property in order to predict the data location
of a sub query. Given dataset D that contains a set
of datasets d, the normalizing number of occurrences
of properties in the Dataset D (OCP(D))is calculated
as follows: OCP(D) =
∑
∀d∈D
|P
d
(d,D)|
|P(D)|×|D|
And the normal-
izing number of occurrences of classes in Dataset D
(OCC(D)) is computed as OCC(D) =
∑
∀d∈D
|C
d
(d,D)|
|C(D)|×|D|
OCP(D) and OCC(D) have a range value from
zero to one. Inspired by the F-Measure function,
we combine OCP(D) and OCC(D) into a single met-
ric which is called the Spreading Factor Γ(D) of the
dataset D. Γ(D) =
(1+β
2
)OCP(D)×OCC(D)
β
2
×OCP(D)+OCC(D)
where β =
0.5
We assign β = 0.5 in order to put more stress on
properties than classes. The intuition is that the high-
est number of the query pattern delivered to SPARQL
endpoint mostly contains constant predicates (Arias
et al., 2011). Moreover, the number of distinct prop-
erties in the dataset is usually higher than the number
of distinct classes in the dataset. The high Γ value
indicates that the class and properties are spread out
over the dataset.
Look back at our previous example in which
we define P
d
(d
1
,D), P
d
(d
2
,D), P(D), C(D),
C
d
(d
1
,D),C
d
(d
2
,D), then we can calculate
OCP(D) =
2+3
3X2
= 0.833 and OCC(D) =
1+1
1X2
= 1.
Finally, we obtain Γ(D) = 1.172
3.2 Spreading Factor of Dataset
associated with the Queryset
The spreading factor of a dataset reveals how the
whole of classes and properties are distributed over
the dataset. However, a query only consists of partial
properties and classes in the dataset. Thus, it is nec-
essary to quantify the spreading factor of the dataset
with respect to the queryset.
Definition 5. A query consists of set of triple patterns
τ which is formally defined as τ(s, p, o) ∈ (U ∪V ) ×
(U ∪V )×(U ∪L∪V ) where V is a set of all variables.
Given a queryset Q = {q
1
,q
2
,· ·· ,q
n
}, the Q-
spreading factor γ of dataset D associated with query-
set Q is computed as γ(Q, D) =
∑
∀q∈Q
∑
∀τ∈q
OC(τ,D)
|Q|
where the occurrences of class and property for τ is
specified as
OC(τ,D) =
o f D(o
τ
,D)
|D|
if p
τ
is rdf:type
∧o
τ
/∈ V
p f D(p
τ
,D)
|D|
if p
τ
is not rdf:type
∧p
τ
/∈ V
∑
∀d∈D
|P
d
(d,D)|
|D|
otherwise
o f D(o,D) denotes the occurrences of object o in
the dataset D and p f D(p, D) denotes the occurrences
of predicate p in the dataset D which can be calculated
as follows: o f D(o,D) =
∑
∀d∈D
o f d(o,d,D) The oc-
currences of object o in the source d can be explained
as follows:
o f d(o,d,D) =
1 if o ∈ C
d
(d,D)
0 otherwise
p f D(p,D) =
∑
∀d∈D
p f d(p,d,D) The occurrence
of predicate p in the source d can be obtained from
the following formula:
p f d(p,d,D) =
1 if p ∈ P
d
(d,D)
0 otherwise
Consider an example, given a query
and a dataset as shown in Figure 1,
then OC(?person a foaf:person,D) = 1 and
OC(?person foaf:name ?name,D) = 1 because
foaf:person and foaf:name are located in two data
sources. As a result, the q-Spreading factor γ(Q, D)
is
1+1
1
= 2
4 EVALUATION
We ran our evaluation on an Intel Xeon CPU X5650,
2.67GHz server with Ubuntu Linux 64-bit installed as
OnMetricsforMeasuringFragmentationofFederationoverSPARQLEndpoints
101

Listing 1: Dailymed Sample Triples.
d a i l y m e d d r u g : 8 2 a d ai l y m e d : d r u g
d a i l y m e d d r u g : 8 2 da i l y m e d : a c t i v e i n g r e d i e n t d a i l y m e d i n g :
P h e n y t o i n
d a i l y m e d d r u g : 8 2 r d f s : l a b e l ” D i l a n t i n −125 ( S u s p e n s i o n ) ”
d a i l y m e d d r u g : 2 0 1 a d a i l ym e d : dr ug
d a i l y m e d d r u g : 2 0 1 d a i ly m e d : a c t i v e i n g r e d i e n t d a i l y m e d i n g :
E t h o s u x i m i d e
d a i l y m e d d r u g : 2 0 1 r d f s : l a b e l ” Z a r o n t i n ( Ca p s u le ) ”
d ai l y m e d o r g : Par ke −Dav is a d a il y m e d : o r g a n i z a t i o n
d ai l y m e d o r g : Par ke −Dav is r d f s : l a b e l ” P ar k e−Dav is ”
d ai l y m e d o r g : Par ke −Dav is da il y m e d : p ro d u c e s Dr ug
d a i l y m e d d r u g : 8 2
d ai l y m e d o r g : Par ke −Dav is da il y m e d : p ro d u c e s Dr ug
d a i l y m e d d r u g : 2 0 1
d a i l y m e d i n g : P h e n y t o i n a d a il y m e d : i n g r e d i e n t s
d a i l y m e d i n g : P h e n y t o i n r d f s : l a b e l ” P h e n y t o i n ”
d a i l y m e d i n g : E th o s u x i m i d e a d a i l ym e d : i n g r e d i e n t s
d a i l y m e d i n g : E th o s u x i m i d e r d f s : l a b e l ” E t h o s u x im i d e ”
the Operating System and Fuseki 1.0 as the SPARQL
Endpoint server. For each dataset, we set up Fuseki
on different ports. We re-used the query set from our
previous work (Rakhmawati and Hausenblas, 2012).
We limited the query processing duration to one hour.
Each query was executed three times on two federa-
tion engines, namely SPLENDID (G
¨
orlitz and Staab,
2011) and DARQ (Quilitz and Leser, 2008). These
engines were chosen because SPLENDID employs
VoID(http://www.w3.org/TR/void/) as data catalogue
that contains a list of predicates and entities, while
DARQ has a list of predicates which is stored in the
Service Description(http://www.w3.org/TR/sparql11-
service-description/). Apart from using VoID,
SPLENDID also sends a SPARQL ASK query to de-
termine whether or not the source can potentially re-
turn the answer. We explain the details of our dataset
generation and metrics as follows:
4.1 Data Distribution
To determine the correlation between the commu-
nication cost of the federated SPARQL query and
the data distribution, we generate 9 datasets by di-
viding the Dailymed(http://wifo5-03.informatik.uni-
mannheim.de/dailymed/) into three partitions based
on following strategies:
4.1.1 Graph Partition
Inspired by data clustering for a single RDF storage
(Huang et al., 2011), we performed graph partition
over our dataset by using METIS (Karypis and Ku-
mar, 1998). The aim of this partition scheme is to
reduce the communication needed between machines
during the query execution process by storing the con-
nected components of the graph in the same machine.
We initially identify the connections of subject and
object in different triples. We only consider the URI
object which is also a subject in other triples. Intu-
itively, the reason is that the object which appears as
the subject in other triples can create a connection if
the triples are located in different dataset partitions.
V (D) denotes the set of pairs of subject and object that
are connected in the dataset D which can be formally
specified as V (D) = {(s, o)|∃s,o, p, p
′
∈ U : (s, p, o) ∈
D ∧ (o, p
′
,o
′
) ∈ D
′
}. We assign a numeric identifier
for each s,o ∈ V (D). After that, we create a list of se-
quential adjacent vertexes for each vertex then uses it
as input of METIS API. Run METIS to divide the ver-
texes and get a list of the partition number of vertexes
as output. Finally, we distribute each triple based on
the partition number of its subject and object. Con-
sider an example, given Listing 1 as a dataset sample,
then
V (D)={(dailymeddrug:82,
dailymeding:Phenytoin),(dailymeddrug:201,
dailymeding:Ethosuximide),(dailymedorg:Parke-Davis,
dailymeddrug:82),(dailymedorg:Parke-Davis,
dailymeddrug:201)}
Starting an identifier value from one and increment
the identifier later, we set the identifier for daily-
meddrug:82 = 1, dailymeding:Phenytoin =2, dai-
lymeddrug:201=3, dailymeding:Ethosuximide=4 and
dailymedorg:Parke-Davis=5. After that, we can
create list of sequential adjacent vertexes V (D) is
{(2,5), 1,(4,5), 3,(1,3)}. Suppose that we divide the
sample of dataset into 2 partitions, then the output of
METIS partition is {1,1,2, 2,1} where each value is
the partition number for each vertex. According to the
METIS output, we can say that dailymeddrug:82 be-
longs to partition 1, dailymeding:Phenytoin belongs
to partition 1, dailymeddrug:201 belongs to partition
2 and so on. In the end, we have two following parti-
tions:
Partition 1: all triples that contain dailymeddrug:82, daily-
meding:Phenytoin and dailymedorg:Parke-Davis
Partition 2: all triples that contain dailymeddrug:201 and
dailymeding:Ethosuximide
4.1.2 Entity Partition
The goal of this partition is to distribute the number of
entities evenly in each partition. Different classes can
be located in a single partition. However, the entities
of the same class should be grouped in the same parti-
tion until the number of entities reaches the maximum
number of entities for each source. We initially create
a list of the subjects along with its class (E(D)). The
set E(D) of pairs of subject and its class in the dataset
D is defined as E(D) = {(s, o)|∃(s,rd f type,o) ∈ D}
WEBIST2014-InternationalConferenceonWebInformationSystemsandTechnologies
102

Then, we sort E(D) by its class o and store each pair
of the subject and object in a partition until the num-
ber of pairs of subject and object equals to the total
pairs of subject and object divided by the number of
partitions. After that, we distribute the remainders of
triples in the dataset based on the subject location.
Given Listing 1 as a dataset sample, then
E(D)={(dailymeddrug:82,dailymed:drug),(dailymeddrug:201
,dailymed:drug),(dailymedorg:Parke-Davis,dailymed:organization),
(dailymeding:Phenytoin,dailymed:ingredients),
(dailymeding:Ethosuximide,dailymed:ingredients)}
Suppose that we split the dataset into two parti-
tions, then the maximum number of entities for each
partition is
|E(D)|
numbero f partitions
=
5
2
= 3 (ceiling 2.5).
We place dailymeddrug:82, dailymeddrug:201 and
dailymedorg:Parke-Davis in the partition 1 and store
the remainders of entities in the partition 2. As the
final step, we distribute the related triples based on its
subject partition number.
4.1.3 Class Partition
Class Partition divides the dataset based on its classes.
The related triples that belong to one entity are placed
in the same machine. To begin with, we also create
E(D) which was used in Entity partition. Later, we
distribute each triple based on the subject class. ike
our previous entity partition example, we do the same
step to generate E(D). However, in the class partition,
we divide the dataset to three partitions since we have
three classes (dailymed:drug, dailymed:organization,
dailymed:ingredients).
4.1.4 Property Partition
Wilkinson(Wilkinson, 2006) introduced a method for
storing RDF data in traditional databases known as
Property Table (PT). There are two types of PT par-
titions: Clustered Property Table and Property-class
Table. In our property partition, we do not have
a Property class table because we treat all proper-
ties in the same manner. We place the triples that
have the same property in one data source. Be-
cause the number of properties in the dataset is gen-
erally high, we allow more than one property to
be stored in the same partition as long as we get
a balanced number of triples among the partitions.
Firstly, we group the triples based on its property.
Next, we store each group in a partition until the
number of partition triples is less than or equal to
the number of dataset triples divided by the num-
ber of partitions. For instance, given a dataset as
shown in Listing 1, then we have four properties:
rdf:type, dailymed:activeingredient, rdf:label and dai-
lymed:producesDrug. Suppose that we want to divide
the dataset into 2 partitions, then the maximum num-
ber of triples in each partition is
thenumbero f triples
thenumbero f partitions
=
14
2
= 7. As the following step, we store the triples
based on its property as follows: Partition 1: five
triples with rdf:type property, two triples with dai-
lymed:activeingredient property and Partition 2: five
triples with rdfs:label property, two triples with daily-
med:producesDrug
4.1.5 Triples Partition
The federation framework performance is influenced
not only by the federated engine solely, but also de-
pends on the SPARQL Endpoints within the federa-
tion framework. In order to keep balanced workload
for SPARQL Endpoints, we split up the triples of each
source evenly because LUBM (Guo et al., 2005) men-
tioned that the number of triples can influence the per-
formance of a RDF repository. We created three triple
partition datasets (T D, T D2, T D3). T D is obtained
by partitioning the native Dailymed dataset into three
parts. T D2 and T D3 are generated by picking a ran-
dom starting point within the Dailymed dump file(by
picking a random line number).
4.1.6 Hybrid Partition
The Hybrid Partition is a partitioning method that
combines two or more previous partition strategies.
For instance, if the number of triples in a class is too
high, we can distribute the triples to another partition
to equalize the number of triples. Since the num-
ber of triples in each dataset of the Class Distribu-
tion CD are not equal, we create HD to distribute
the triples evenly. However, rdf:type property and
rdfs:label property are evenly through all partitions in
dataset HD2. This distribution is intended for balanc-
ing the workload amongst SPARQL Endpoints since
those properties are commonly used in our query set.
As shown in those figures, the classes and proper-
ties are distributed over most of the partitions in the
GD dataset. The PD has the lowest Spreading Fac-
tor among the dataset because each property occurs
in exactly one partition and only in one partition has a
set of triples that contains rdf:type. The dataset gen-
eration code and the generation results can be found
at DFedQ github(https://github.com/nurainir/DFedQ)
4.2 Metrics
To calculate the communication cost of the the fed-
erated SPARQL query, we compute the data transfer
OnMetricsforMeasuringFragmentationofFederationoverSPARQLEndpoints
103

PD CD TD TD3 TD2 ED HD2 HD GD
Dataset Partitions
.000
.400
.800
1.200
Spreading Factor of The datasets
Figure 2: Spreading Factor of Dataset.
volume between the federated engine and SPARQL
Endpoints. The data transfer volume includes the
amount of data both sent and received by the me-
diator. Apart from capturing the data transmission,
we also measure the requests workload (RW ) during
query execution. RW is calculated as RW =
RQ
T ∗SS
where RQ refers to the number of requests sent by
the federated engine to all SPARQL Endpoints, T de-
notes the duration between when a query is received
by the federated engine and when its results starts to
be dispatched to the client and SS is the number of
selected sources. Furthermore, we also measure the
response time that is required by a federated engine
to execute a query.
For the sake of readability, we aggregate each per-
formance metric results into a single value. In or-
der to avoid trade-offs among queries, we assign a
weight to each query using the the variable counting
strategy from the ARQ Jena (Stocker and Seaborne,
2007). This weight indicate the complexity of the
query based on the selectivity of the variable posi-
tion and the impact of variables on the source selec-
tion process. The complexity of query can influence
the federation performance. Hence, we normalize
each performance metric result by dividing the met-
ric value with the weight of the associated query. In
the context of federated SPARQL queries, we set the
weight of the predicate variable equals to the weight
of the subject variable since most of the federated en-
gines rely on a list of predicates to decide the data
location. Note that, a triple pattern can contain more
than one variable. The details of the weight of subject
variable w
s
, predicate variable w
p
and object variable
w
o
for the triple pattern τ can be explained as follows:
w
s
(τ)=
3 if the subject of triple pattern τ ∈ V
0 otherwise
w
p
(τ)=
3 if the predicate of triple pattern τ ∈ V
0 otherwise
w
o
(τ)=
1 if the object of triple pattern τ ∈ V
0 otherwise
Finally, we can compute the weight of query
q: weight(q) =
∑
∀τ∈q
w
s
(τ)+w
p
(τ)+w
o
(τ)+1
MAX COST
where
MAX COST = 8 because if a triple pattern consists of
variables that are located in all positions, the weight of
the triple pattern is 8(3+3+1+1). By using the weight
of a query, we can align the query performance re-
sults afterwards. We do not create a composite metric
that combines the response time, the request workload
and the data transfer, but rather we calculate each per-
formance metric results individually. Given that Q is
a set of queries q in the evaluation and that m is a
set of performance metric results associated with the
queryset Q, then the final metric µ for the evaluation
is µ(Q, m) =
∑
∀q∈Q
m
q
weight(q)
|Q|
For instances, the query in Figure 1 has a weight
=
3+1
8
+
3+1+1
8
= 1.125. Suppose that the volume of
data transmission during this query execution is 10
Mb and we only have one query in the queryset, then
µ(Q,m) can be calculated
10
1.125
1
= 8.88Mb.
5 RESULTS AND DISCUSSION
As seen in Figures 3 and 4, the data transmission
between DARQ and SPARQL Endpoints is higher
than the data transmission between SPLENDID and
SPARQL Endpoints. However, Figures 5 and 6 show
that the average requests workload in DARQ is less
than the average requests workload in SPLENDID.
This is because DARQ never sends SPARQL ASK
queries in order to predict the most relevant source
for each sub query.
Overall, data transmission increases gradually in
line with the Spreading Factor of a dataset. However,
the data transmission rises dramatically for GD dis-
tribution. This indicates that in the context of Fed-
erated SPARQL queries, data clustering based on its
property and class is better than data clustering based
on related entities such as Graph Partition. The rea-
son behind this conclusion is that the source selec-
tion in federated query engine depends on classes and
properties occurrences. Furthermore, when the feder-
ated engines generate query plans, they use optimiza-
tion techniques based on the statistical predicates and
classes.
Although a small Spreading Factor can mini-
mize the communication cost, it can also reduce the
SPARQL Endpoint performance. As shown in Fig-
ure 5 and 6, a small Spreading Factor can lead to
the high number of requests received by SPARQL
Endpoint in one second because in the property dis-
tribution, the federated engine mostly sends differ-
ent query patterns to multiple datasource. More-
over, the SPARQL endpoint that stores the popular
predicates such as rdf:type and rdfs:label will receive
WEBIST2014-InternationalConferenceonWebInformationSystemsandTechnologies
104
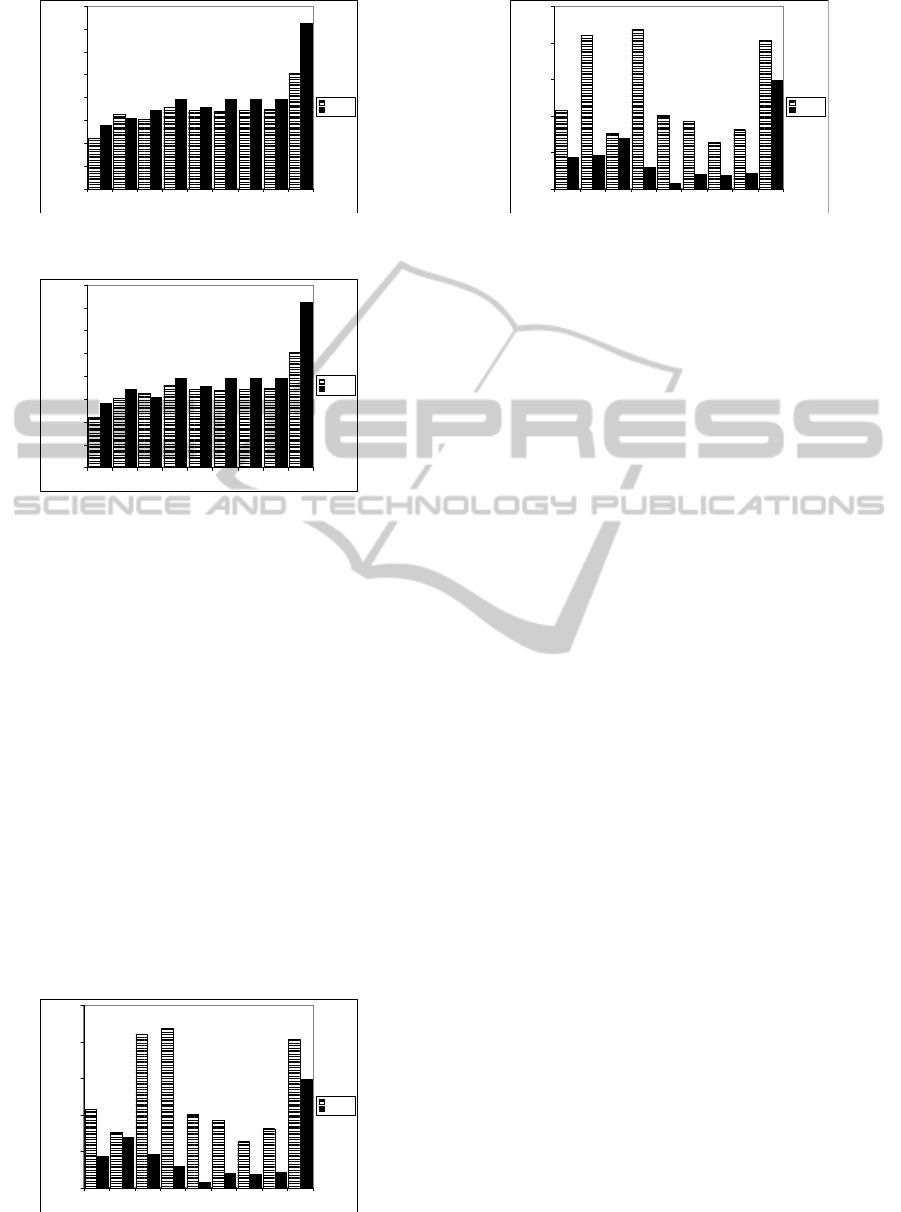
PD CD TD TD3 TD2 ED HD2 HD GD
Dataset Partitions
0E+00
500E+03
1E+06
2E+06
2E+06
2E+06
3E+06
4E+06
4E+06
Average Data Transmission (Bytes)
Splendid
DARQ
Figure 3: Average Data Transfer Volume Vs the Spreading
Factor of Datasets (order by the Spreading Factor value).
PD TD CD TD3 TD2 ED HD2 HD GD
Dataset Partitions
0E+00
500E+03
1E+06
2E+06
2E+06
2E+06
3E+06
4E+06
4E+06
Average Data Transmission (Bytes)
Splendid
DARQ
Figure 4: Average Data Transfer Volume Vs the Q-
Spreading Factor of Datasets associated with the Query-
set(order by the Spreading Factor value).
more requests than other SPARQL endpoints. Con-
sequently, this such condition can lead to incomplete
results because when overloaded, the SPARQL End-
point might reject requests (e.g Sindice SPARQL end-
point(http://sindice.com/) only allows one client send-
ing one query per second). Poor performance is also
shown at the highest value of Spreading Factor of
the dataset (GD) because the entities are spread over
the dataset partitions. Hence, with the calculation of
the spreading factor of the dataset, the federated en-
gine can create a query optimization which attempts
to adapt the dataset characteristic that is shown from
the spreading factor value. For instance, if the dataset
has too small Spreading Factor, the federated engine
should maintain a timer to send several requests to the
same SPARQL endpoint in order to keep the sustain-
ability of the SPARQL endpoint as well as avoid the
PD CD TD TD3 TD2 ED HD2 HD GD
Dataset Partitions
0E+00
5E+00
10E+00
15E+00
20E+00
25E+00
Average Requests Workload (Requests/Second)
Splendid
DARQ
Figure 5: Average Requests Workload Vs the Spreading
Factor of Datasets(order by the Spreading Factor value).
PD TD CD TD3 TD2 ED HD2 HD GD
Dataset Partitions
0E+00
5E+00
10E+00
15E+00
20E+00
25E+00
Average Requests Workload (Requests/Second)
Splendid
DARQ
Figure 6: Average Requests Workload Vs the Q-Spreading
Factor of Datasets associated with the Queryset(order by the
Spreading Factor value).
incomplete answer.
6 CONCLUSION
We have implemented various data distribution strate-
gies to partition classes and properties over dataset
partitions. We introduced two notions of dataset met-
rics, namely the Spreading Factor of a dataset and
the Spreading Factor of a Dataset associated with the
query set. These metrics expose the distribution of
classes and properties over the dataset partitions. Our
experiment results revealed that the class and property
distribution effects on the communication cost be-
tween the federated engine and SPARQL endpoints.
However, it does not significantly influence the re-
quest workload of a SPARQL endpoint. Partitioning
triples based on the properties and classes can mini-
mize the communication cost. However, such parti-
tioning can also reduce the performance of SPARQL
endpoints within the federation infrastructure. Fur-
ther, it can also influence the overall performance of
federation framework.
In future work, we will apply other dataset par-
titioning strategies and use more federated query
engines which have different characteristics from
DARQ and SPLENDID.
ACKNOWLEDGEMENTS
This publication has emanated from research con-
ducted with the financial support of Science
Foundation Ireland (SFI) under Grant Number
SFI/12/RC/2289 and Indonesian Directorate General
of Higher Education. Thanks to Soheila for a great
discussion
OnMetricsforMeasuringFragmentationofFederationoverSPARQLEndpoints
105

REFERENCES
Abadi, D. J., Marcus, A., Madden, S. R., and Hollenbach,
K. (2007). Scalable semantic web data management
using vertical partitioning. In Proceedings of the 33rd
international conference on Very large data bases,
VLDB ’07, pages 411–422.
Arias, M., Fern
´
andez, J. D., Mart
´
ınez-Prieto, M. A., and
de la Fuente, P. (2011). An empirical study of real-
world sparql queries. CoRR, abs/1103.5043.
Duan, S., Kementsietsidis, A., Srinivas, K., and Udrea,
O. (2011). Apples and oranges: a comparison of
rdf benchmarks and real rdf datasets. In ACM Inter-
national Conference on Management of Data (SIG-
MOD).
G
¨
orlitz, O. and Staab, S. (2011). SPLENDID: SPARQL
Endpoint Federation Exploiting VOID Descriptions.
In COLD2011, Bonn, Germany.
Guo, Y., Pan, Z., and Heflin, J. (2005). Lubm: A bench-
mark for owl knowledge base systems. Web Seman-
tics: Science, Services and Agents on the World Wide
Web, 3(2-3):158 – 182.
Huang, J., Abadi, D. J., and Ren, K. (2011). Scalable sparql
querying of large rdf graphs. PVLDB, 4(11):1123–
1134.
Karypis, G. and Kumar, V. (1998). A fast and high qual-
ity multilevel scheme for partitioning irregular graphs.
SIAM J. Sci. Comput., 20(1):359–392.
Montoya, G., Vidal, M.-E., Corcho,
´
O., Ruckhaus, E., and
Aranda, C. B. (2012). Benchmarking federated sparql
query engines: Are existing testbeds enough? In In-
ternational Semantic Web Conference (2), pages 313–
324.
Prasser, F., Kemper, A., and Kuhn, K. A. (2012). Ef-
ficient distributed query processing for autonomous
rdf databases. In Proceedings of the 15th Interna-
tional Conference on Extending Database Technol-
ogy, EDBT ’12, pages 372–383, New York, NY, USA.
ACM.
Quilitz, B. and Leser, U. (2008). Querying distributed rdf
data sources with sparql. In ESWC2008, pages 524–
538, Berlin, Heidelberg. Springer-Verlag.
Rakhmawati, N. A. and Hausenblas, M. (2012). On the im-
pact of data distribution in federated sparql queries. In
Semantic Computing (ICSC), 2012 IEEE Sixth Inter-
national Conference on, pages 255 –260.
Rakhmawati, N. A., Umbrich, J., Karnstedt, M., Hasnain,
A., and Hausenblas, M. (2013). Querying over feder-
ated sparql endpoints - a state of the art survey. CoRR,
abs/1306.1723.
Schmidt, M., G
¨
orlitz, O., Haase, P., Ladwig, G., Schwarte,
A., and Tran, T. (2011). Fedbench: A benchmark suite
for federated semantic data query processing. In ISWC
Schmidt, M., Hornung, T., Lausen, G., and Pinkel, C.
(2009). Spˆ 2bench: a sparql performance benchmark.
In Data Engineering, 2009. ICDE’09. IEEE 25th In-
ternational Conference on, pages 222–233. IEEE.
Schwarte, A., Haase, P., Schmidt, M., Hose, K., and
Schenkel, R. (2012). An experience report of large
scale federations. CoRR, abs/1210.5403.
Stocker, M. and Seaborne, A. (2007). Arqo: The architec-
ture for an arq static query optimizer.
Wilkinson, K. (2006). Jena property table implementation.
In In SSWS.
WEBIST2014-InternationalConferenceonWebInformationSystemsandTechnologies
106
