
A Multiagent-based Framework for Solving Computationally
Intensive Problems on Heterogeneous Architectures
Bioinformatics Algorithms as a Case Study
H. M. Faheem
1
and B. König-Ries
2
1
Computer Systems Department, Faculty of Computer and Information Sciences, Ain Shams University, Cairo, Egypt
2
Faculty of Mathematics and Computer Science, Jena University, Jena, Germany
Keywords: Bioinformatics, Heterogeneous Architectures, Motif Finding Problem and Multiagent Systems.
Abstract: The exponential increase of the amount of data available in several domains and the need for processing
such data makes problems become computationally intensive. Consequently, it is infeasible to carry out
sequential analysis, so the need for parallel processing. Over the last few years, the widespread deployment
of multicore architectures, accelerators, grids, clusters, and other powerful architectures such as FPGAs and
ASICs has encouraged researchers to write parallel algorithms using available parallel computing paradigms
to solve such problems. The major challenge now is to take advantage of these architectures irrespective of
their heterogeneity. This is due to the fact that designing an execution model that can unify all computing
resources is still very difficult. Moreover, scheduling tasks to run efficiently on heterogeneous architectures
still needs a lot of research. Existing solutions tend to focus on individual architectures or deal with
heterogeneity among CPUs and GPUs only, but in reality, often, heterogeneous systems exist. Up to now
very cumbersome, manual adaption is required to take advantage of these heterogeneous architectures. The
aim of this paper is to provide a proposal for a functional-level design of a multiagent-based framework to
deal with the heterogeneity of hardware architectures and parallel computing paradigms deployed to solve
those problems. Bioinformatics will be selected as a case study.
1 INTRODUCTION
Heterogeneous architectures in modern data centers
include different subsystems that may have CPUs,
GPUs, grids, clusters, FPGAs, and ASICs.
Performance of each subsystem in handling
computationally intensive problems depends mainly
on its computing power. Unified access to all the
heterogeneous systems is still in its initial phases.
Several trials to solve the heterogeneity among
CPUs and GPUs are currently available (Augonnet,
C., et. al, 2009), (Arabnejad, H. and Barbosa, J.,
2013), and (Rauber and Rünger, 2010). These trials
offered run-time systems that allowed the
programmer to select or even provide a user-defined
scheduling strategy but they didn’t provide any
support to FPGA, ASIC, or any other special
purpose parallel architectures. Other trials are
focusing on CPUs, GPUs, and FPGAs (Inta, R.,
Bowman, D., and Scott, M., 2012). These trials are
proposing the design of algorithms that can use all
the existing resources in the machine or a cluster
such that the algorithm will manipulate the CPU,
GPU, and FPGA. Algorithms design is completely
depending on the programmer capabilities in
allocating the hardware resources efficiently.
However, all the trials didn’t provide any
mechanism to automatically schedule tasks
according to the existing hardware. Currently,
neither standards nor functional-level descriptions
are available to define necessary rules or functions
to efficiently schedule tasks on heterogeneous
architectures. In principle, having a framework that
is able to integrate different heterogeneous
architectures and treating them as a unified
computing resource constitutes a dream to
programmers. The intended framework should be
able to: 1) interactively analyze the task dependency
of the algorithm used to solve a given problem, 2)
dynamically allocate computing resources with
tasks, 3) autonomously respond to hardware
topology changes, and 4) intelligently generate
relevant parallel codes that best fit the existing
computing resources. The software paradigm best
526
M. Faheem H. and König-Ries B..
A Multiagent-based Framework for Solving Computationally Intensive Problems on Heterogeneous Architectures - Bioinformatics Algorithms as a Case
Study.
DOI: 10.5220/0004967105260533
In Proceedings of the 16th International Conference on Enterprise Information Systems (ICEIS-2014), pages 526-533
ISBN: 978-989-758-027-7
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)
fitting these requirements is the multiagent-based
system (Russel and Norvig, 1995). What we are
proposing in this paper is a functional-level design
of automatic but intelligent task scheduling
mechanism that distributes tasks after exploring the
existing hardware and then allows for automatic
parallel code generation (using available parallel
computing paradigms) for each hardware resource.
This will be carried out using a multiagent-based
framework. The remainder of this paper is organized
as follows: Section 2 explores the suggested
multiagent-based framework. Section 3 describes
how the four layers can be customized to suit the
bioinformatics domain. Section 4 illustrates the
structure of the multiagent-based framework. It also
provides some attributes of the agents such as name,
percept sequence, action, goals, and type. Section 5
shows how the framework can be used to solve the
motif finding problem as an example. Section 6
provides conclusion and directions for future work.
2 MULTIAGENT-BASED
FRAMEWORK FOR SOLVING
COMPUTATIONALLY
INTENSIVE PROBLEMS
The suggested multiagent framework for solving
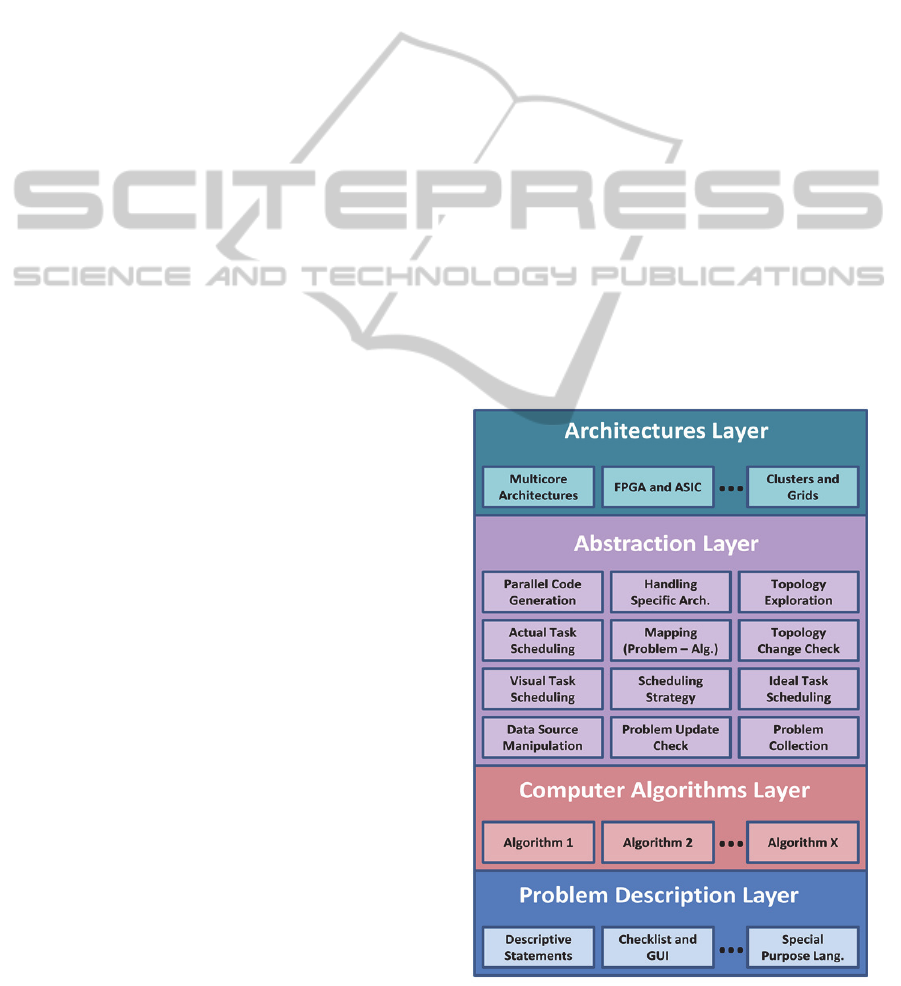
computationally intensive problems depicted in Fig.
1 consists of four layers: problem description layer,
computer algorithms layer, abstraction layer, and
architectures layer. The problem description layer
describes the problem. The description of the
problem can be provided in several ways. It can be
presented as a checklist, written in a special purpose
language, or simply written as a set of descriptive
statements. Checklists allow the agent program to
automatically generate the relevant actions. In case
of a written problem with a special purpose
language, a compiler is needed to compile the
language statements and produce necessary
executable codes. Descriptive statements need much
more effort since domain expert intervention is
required to pass the parameters, attributes, and
formulations to the system. The computer
algorithms layer is responsible for mapping the
problem from its specific domain (bioinformatics,
climatology, etc.) to the computer domain. It is
interested in the use of computer and information
sciences and mathematics to model and analyze the
problem. The abstraction layer contains intelligent
agents responsible for performing several tasks
among them: gathering computer algorithms of a
specific problem, exploring architectures features,
checking for updates of problems, checking for
updates of hardware architectures features, ideal
scheduling of tasks, setting scheduling strategy,
actual scheduling of tasks, visual presentation of
task dependency diagrams, and managing the
configuration of hardware architectures using the
drivers of these architectures. Agents can traverse
across different layers to perform their intended
tasks. This layer also provides the mapping between
the algorithm intended to solve the problem and the
intended hardware architectures to perform this
algorithm. This layer constitutes the shielding of
problem domain researchers from hardware
architectures level configuration details. The
architectures layer contains different hardware
architectures that can communicate and cooperate
with agents. Software drivers of architectures can
negotiate with the system agents such that data can
be sent and received by the system agents. The
software drivers should support different
architecture requirements such as programs and data
handling mechanisms. ASIC, FPGA, and DNA-
based Self Assembled Architectures should have
their own drivers developed by the hardware
manufacturer and should be able to accept
configuration profiles or scripts. Clusters and other
Figure 1: Layers of the Multiagent-based Framework.
AMultiagent-basedFrameworkforSolvingComputationallyIntensiveProblemsonHeterogeneousArchitectures-
BioinformaticsAlgorithmsasaCaseStudy
527
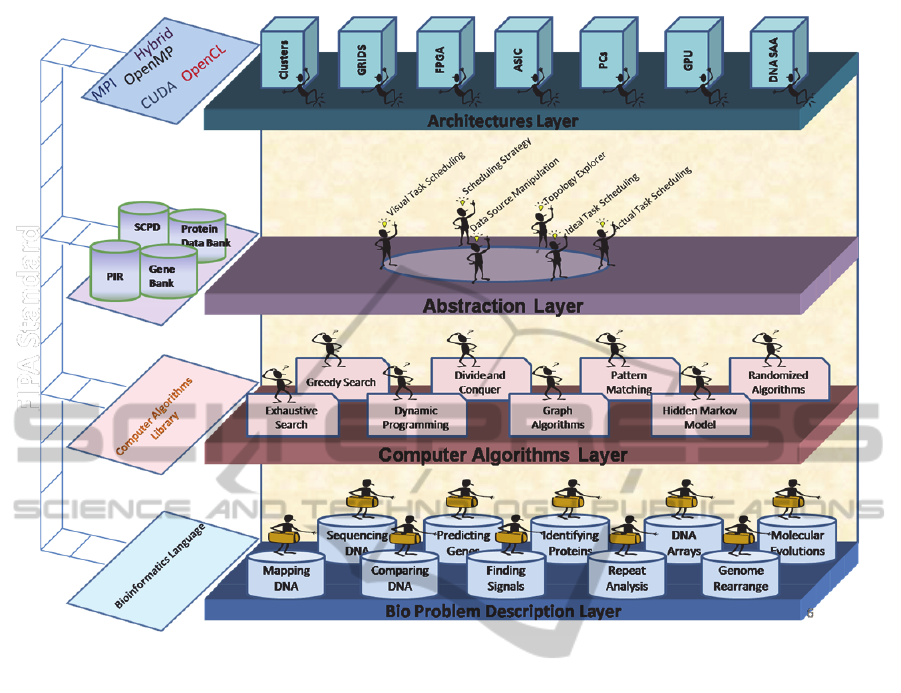
Figure 2: The Four Layers of Multiagent-based Framework for Bioinformatics.
standard systems can communicate directly with the
multiagent system using standard multiagent
schemes.
3 MAPPING THE FOUR LAYERS
TO THE BIOINFORMATICS
DOMAIN
Hugh amounts of biological data deposited in Web
databases are currently available. Access to this data
is very important to biological researchers.
Accessibility to such databases has encountered a lot
of difficulties due to the heterogeneity among
biological databases in data formats, data
representations, and data source schema (Miled et
al., 2003).
Bioinformatics tools proved remarkable success
in different areas of bioinformatics like gene finding
and sequence alignment. Many approaches have
been proposed and one can find many published
papers describing novel algorithms to address such
computationally intensive bioinformatics problems.
Parallelism seems to be the trend. Different
bioinformatics algorithms are currently developed in
a parallel format such that a significant improvement
in terms of speedup has been achieved. Parallel
processing researchers are thinking of the
Bioinformatics problems from the point of studying
task dependency such that concurrent execution of
parallel tasks can dramatically reduce the overall
execution time required to solve a given problem.
Biologists are thinking of the problem in a different
way such that they believe they have to find
relations and correlations related to different
sequences irrespective of the time constraint. They
spend a lot of time seeking for solutions to their
computationally intensive problems. In fact all they
need is a simple way to issue an order to find a
certain motif, or align a sequence, etc. such that they
can proceed in their work to extract conclusions.
Bioinformatics researchers are trying to understand
the problem definition from the biologists in order to
invent new algorithms to solve such given problems.
In doing so, they are trying to convert the problem
from the biology domain to the information
processing domain. A set of questions may arise,
ICEIS2014-16thInternationalConferenceonEnterpriseInformationSystems
528

among them:
Can we develop a simple bioinformatics
language that can be used by researchers of
biology to simply perform a certain task or solve
a specific problem?
Can we design a system that is able to identify
the problem and extract the implicit parallelism,
study task dependency, and provide an ideal task
scheduling mechanism?
Can we query the existing and available
hardware infrastructure in a way that can guide
us to map the tasks efficiently to be implemented
on such hardware platforms irrespective of their
heterogeneity?
Can we perform these tasks in an intelligent
way?
Several trials have been concerned with deploying
agent-oriented technology in the Bioinformatics
domain. Most of them were focusing on integrating
biological data from different data sources as in
(Shunmaganathan, Deepika, and Deeba, 2008).
Agent-oriented technology has also been deployed
in task scheduling as in (Konwinski, 2012). In our
proposal we show how to deal with the
heterogeneity of data sources, hardware
architectures, and parallel computing paradigms
deployed to solve bioinformatics problems.
The advent of agent technology for
bioinformatics yields remarkable advantages. Since
the multiagent system deploys a concurrent,
cooperative working technique then it can fit easily
to the distributed programming approach used for
parallel bioinformatics algorithms. A multiagent
system consists of several agents that can run on a
distributed system. Agents can cooperate with each
other to perform a specific task or a set of tasks.
During parallel processing of a bioinformatics
algorithm, a set of processing units work in a
distributed system and communicate via data
streams to perform tasks. This conforms well with
decomposition of processing tasks to dedicated
agents which in turn coordinate and perform tasks.
Agents are appropriate for efficient, distributed
planning. This encourages the utilization of agents
for planning the parallel processing of
bioinformatics tasks. Agents can be used for
distributed resource management. Consequently,
they will be used for distributing data and collecting
results. Agents are able to plan and perform parallel
bioinformatics algorithms using parallel processing.
Rational agents have the ability of learning. This
enables them to implement dynamic load balance
strategies that can be used to handle optimal
distribution of data and tasks. Moreover, the ability
of intelligent agents to learn can help in improving
the scheduling strategies of the agent. The four
layers of the proposed multiagent-based system can
be related to the bioinformatics domain as shown in
Fig. 2. A set of bioinformatics problems is presented
in the problem description layer. Bioinformatics
algorithms are included in computer algorithms
layer. Abstraction layer has agents that perform
some functions described earlier. Some agents from
this layer can traverse to other layers such that they
can visit the problem layer to collect the problem,
visit the architectures layer to collect hardware
features, and can also visit the computer algorithms
layer to select the appropriate algorithm for solving
a given bio problem. The fourth layer
“Architectures Layer” contains the same set of
heterogeneous hardware architectures.
4 MULTIAGENT-BASED
FRAMEWORK STRUCTURE
A bioinformatics problem is picked up from a text
file written by the biologist using the Bio Problem
Collector Agent. This agent is responsible for
interpreting the biology problem into a specific
bioinformatics language. The agent can also provide
a GUI to allow the bioinformatics programs
developer to simply select a suitable set of
statements. It is assumed that the problem writer has
enough experience to select among different
bioinformatics language statements. The bio
problem collector agent periodically collects bio
problems and forwards them to the Mapping Agent.
The Bio Problem Updater Agent tracks changes and
searches for any updates into the problem and
provides these updates to the Mapping Agent that
specifies the class of the computer algorithms
relevant to the bio problem and then forwards this
classification to the Ideal Task Scheduling Agent.
The Ideal Task Scheduling Agent is responsible for
analyzing the problem and extracting the task
dependency diagram irrespective of the hardware
architecture it will run on. The task dependency
diagram generated by the Ideal task scheduling
agent is forwarded to the Scheduling Strategy Agent.
The Feature Collector Agent provides the Topology
Explorer Agent with specific details of the hardware
architecture it belongs to. The Topology Explorer
Agent collects different architectures features and
status and forwards them to the Scheduling Strategy
Agent. Now the Scheduling Strategy Agent has both
the task dependency diagram (tree) and the available
hardware topology and features. The Scheduling
AMultiagent-basedFrameworkforSolvingComputationallyIntensiveProblemsonHeterogeneousArchitectures-
BioinformaticsAlgorithmsasaCaseStudy
529
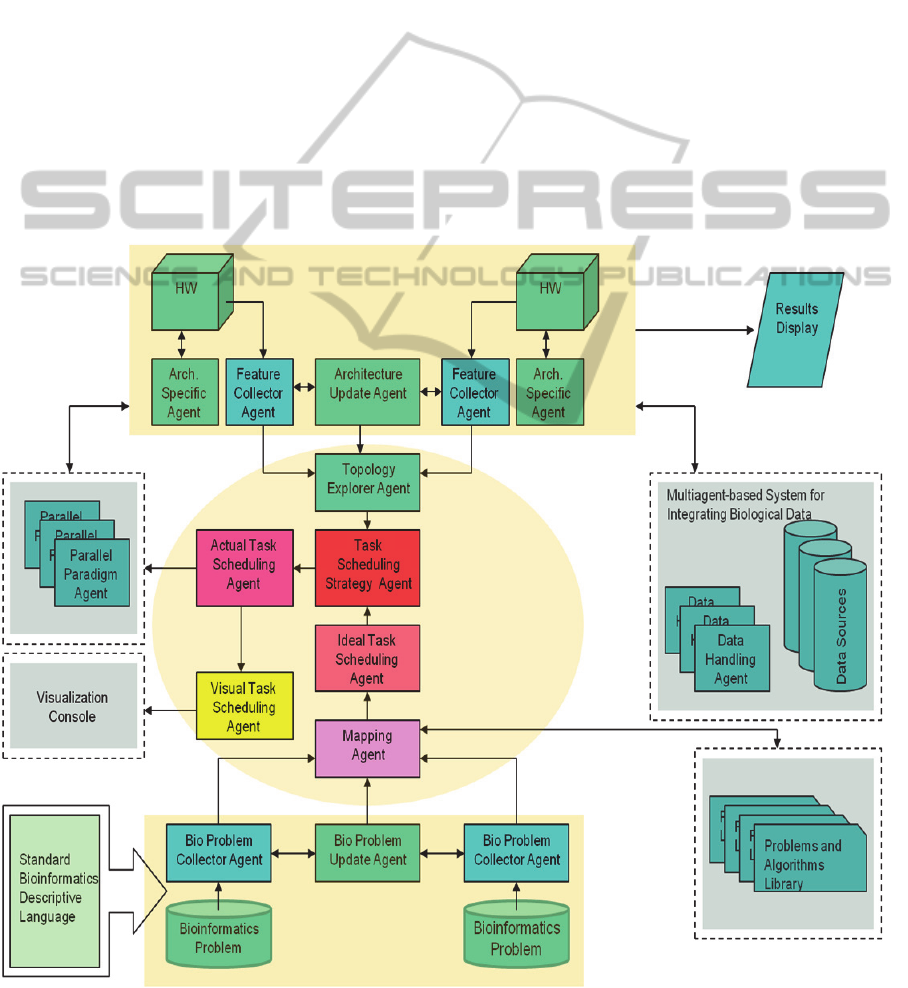
Strategy Agent is now able to decide on the
appropriate scheduling strategy that best assigns
tasks to hardware architectures. The Actual Task
Scheduling Agent enforces the scheduling strategy
and assigns specific tasks to specific hardware
architectures and sends this assignment to the
Parallel Paradigm Agent(s) which in turn generates
the relevant code that will run on the relevant
architectures. The code generated by the Parallel
Paradigm Agent(s) will then move to the
Architecture Specific Agent which is responsible for
dealing with its specific architecture. The
Architecture Update Agent keeps track of any
changes related to the hardware architecture and
forwards these changes to the Topology Explorer
Agent. In fact, some sort of integration with a
multiagent-based system for integrating biological
data should be addressed. Data coming from
different data sources to be entered to the hardware
architectures for executing the intended code should
come through standard and common agent interface
as in (Maghrabi, F., et al, 2008). Other agents may
perform some task monitoring functions such as
Visual Task Scheduling Agent. A brief description of
each agent including its name, percept sequence,
actions, goals, and type is listed in Table 1 while the
suggested multiagent-based framework structure is
shown in Fig.3
5 SOLVING MOTIF FINDING
PROBLEM
Motif is generally defined as a recurring pattern in
the sequence of nucleotides or amino acids. In the
DNA sequence, it is usually a short segment that
occurs frequently, but not required to be an exact
copy for each occurrence (typical pattern matching
problem). The Motif Finding Problem MFP has
Figure 3: Multiagent-based System Structure.
ICEIS2014-16thInternationalConferenceonEnterpriseInformationSystems
530
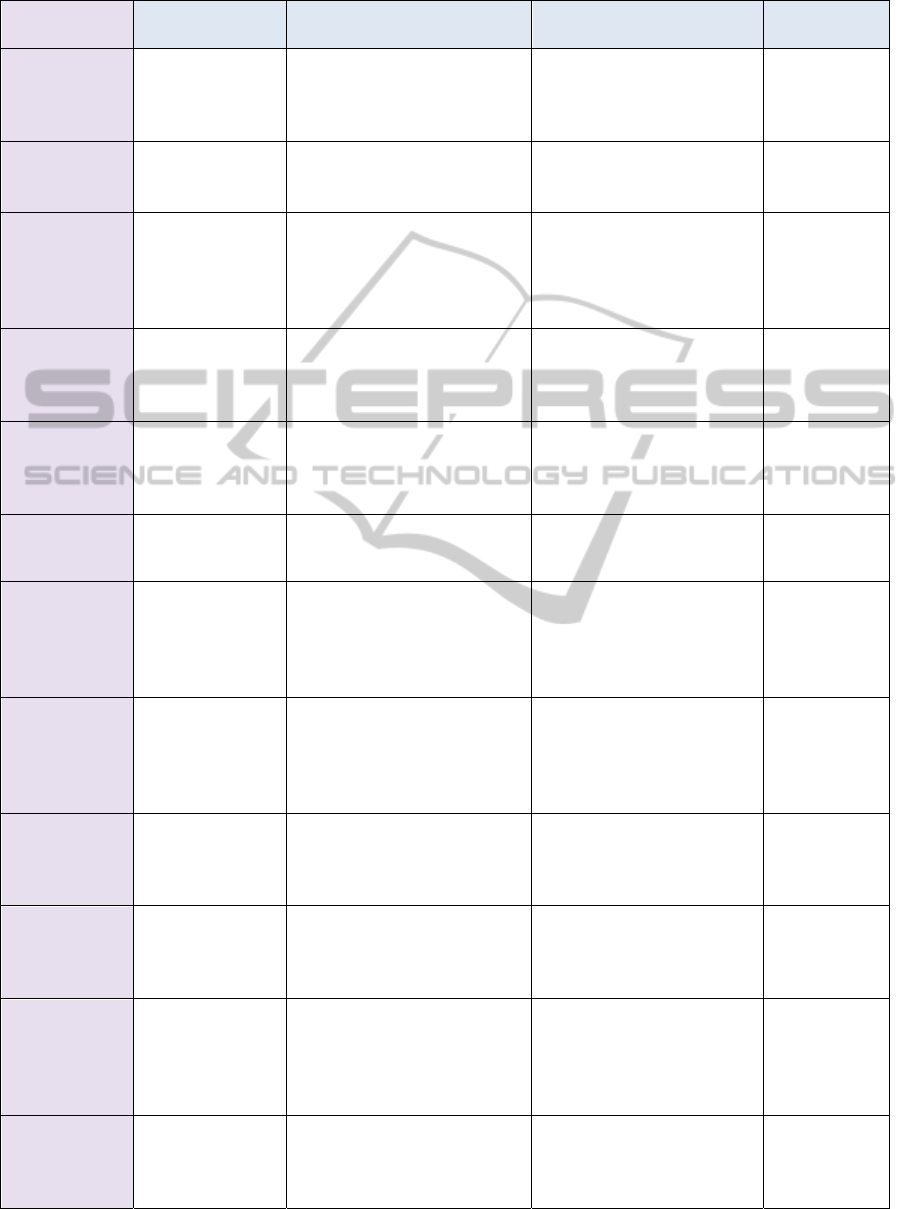
Table 1: Agents Description.
Agent Name Percepts Actions Goals Type
Bio Problem
Collector
Written Bio
Problem
Mapping problem into
standard common bio-
problem (written in
bioinformatics language)
Keep track of the bio
problem
Simple Reflex
Bio Problem
Updater
Written Bio
Problem
Matches gathered problem
with previously stored
problem
Notification of Mapping
Agent with new problem
updates
Agent that
keeps track of
the world
Mapping
Bio Problems
and updates
(written in
Bioinformatics
language)
Classifies received bio
problem and then maps it to
an equivalent computer
algorithm
Determination of the
matched class of the
computer algorithm
Goal-based
Ideal Task
Scheduling
Computer
Algorithm
describing the
bio problem
Analyze the algorithm to
extract task dependency
diagram
Extracting task
dependencies of the
algorithm
Utility-based
Task
Scheduling
Strategy
Ideal task
scheduling and
available
architectures
Analysis of the properties of
different architectures to
allocate a certain set of tasks
to each
Deciding the appropriate
scheduling strategy that
best fit the available
architectures
Utility-based
Actual Task
Scheduling
Strategy of
scheduling
Applying scheduling
strategy
Task distribution to each
architecture
Utility-based
Topology
Explorer
Receives
different features
of the available
architectures or
features updates
List available architectures,
its status, its attributes, and
suitable computing paradigm
Clarify the available
topology structure
Utility-based
Architecture
Specific
Instructions from
Parallel Paradigm
Agent
Converts parallel
instructions to scripts and
negotiates drivers of
architectures to perform
necessary operations
Setting up, configuring,
and running appropriate
code on a specific
architecture
Utility-based
Feature
Collector
Specific
architecture
features
Mapping the gathered
architecture features into a
readable format suitable for
the topology explorer agent
Keep track of the
architecture features
Simple Reflex
Architecture
Updates
Architecture
Features
Forwarding architecture
features or updates to
Topology Explorer Agent
Notification of the
Topology Explorer Agent
with new updates of the
architecture
Agent that
keeps track of
the world
Parallel
Paradigm
Actual task
scheduling
scheme
Converting actual task
scheduling scheme into a
well-defined code that can
run on a specific architecture
Submitting a specific code
written in a specific
parallel paradigm to an
Architecture Specific
Agent
Utility-based
Visual Task
Scheduling
Actual task
scheduling
scheme
Convert the actual task
scheduling into a graphical
diagram showing task
dependencies
Providing a visual
monitoring for task
dependencies and their tree
Goal-based
AMultiagent-basedFrameworkforSolvingComputationallyIntensiveProblemsonHeterogeneousArchitectures-
BioinformaticsAlgorithmsasaCaseStudy
531
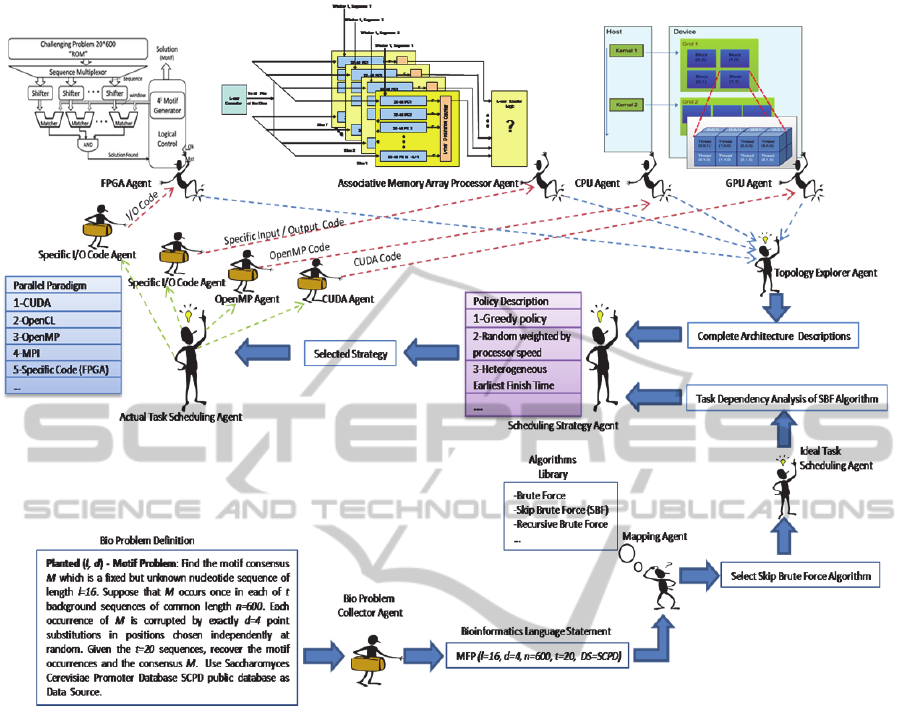
Figure 4: The operation of the multiagent-based framework to solve motif finding problem.
been tackled several times as in (Rajasekaran, Balla,
Huang, 2005). Different architectures and algorithms
are designed to solve such computationally intensive
problem. MFP can be illustrated as follows: Planted
(l, d) - Motif Problem: Find the motif consensus M
which is a fixed but unknown nucleotide sequence
of length l. Suppose that M occurs once in each of t
background sequences of common length n. Each
occurrence of M is corrupted by exactly d point
substitutions in positions chosen independently at
random. Given the t sequences, recover the motif
occurrences and the consensus M. We also will
consider that we have a typical
heterogeneous environment having CPUs, GPUs,
FPGA architecture as in (Farouk, El-Deeb, and
Faheem, 2011), and ASIC as in (Faheem, 2010). The
skip brute force SBF algorithm was selected to solve
the MFP. The operation of the multiagent-based
framework is illustrated in Fig. 4. It is well
understood that there is no task dependency after
expanding input sequences since comparison
processes are carried out between a specific l-mer
and all the input sequences windows that have the
same length. It is assumed that we will use
Saccharomyces Cerevisiae Promoter Database
SCPD public database as a data source. This is
obvious in the statement generated by the bio
problem collector agent.
The skip brute force SBF algorithm has been
selected by the mapping agent to solve the MFP.
Each agent in the proposed framework performs a
specific operation to perform a specific task to solve
the problem of MFP. However, special focus on the
task scheduling strategy agent should be taken into
account. This agent decides the scheduling strategy
based on the topology of the heterogeneous
architectures and the set of tasks to be performed.
Predefined scheduling strategies should be supported
such as greedy policy, priority queues,
Heterogeneous Earliest Finish Time, etc. User-
defined policies should also be supported such that
the user can define his scheduling policy.
ICEIS2014-16thInternationalConferenceonEnterpriseInformationSystems
532

The actual task scheduling agent in our case will
provide the parallel paradigm agents with the actual
task list to be executed. Clearly, CUDA agent will
generate the appropriate code for the GPU.
OpenMP agent will generate the appropriate code
for the multicore CPUs. Specific Input / Output code
will be generated to allow data exchange between
the FPGA agent and its relevant hardware and the
same concept will be applied to the associative
memory array processor.
6 CONCLUSION
In our trial to standardize the functions related to
solving computationally intensive problems on
heterogeneous architectures, a functional level
description of a multiagent-based framework is
proposed. The bioinformatics domain has been
selected as a case study. The function of each agent
in the system is clarified. The operation of the
system is described through an example of solving
MFP. The framework is in its initial phase. As a
next step, the actual development of such proposed
system will be implemented using available
multiagent based frameworks such as JADE,
EtherYatri, AgentBuilder, etc. Clarification of the
initial rules that will be used by each agent will be
addressed. Learning mechanisms of the agents will
also be considered. We believe that it is an initial
draft version of a multiagent-based system that can
be established and can move towards an efficient
system to solve computationally intensive problems
on heterogeneous architectures.
REFERENCES
Miled, Z. et al., 2003. An Ontology for Semantic
Integration of Life Science Web Databases.
International Journal of Cooperative Information
Systems.12 (02).
Rauber, T., Rünger, G., 2010.Parallel Programming: for
Multicore and Cluster Systems.Springer.
Farouk, Y., El-Deeb, T., and Faheem, H., 2011.Massively
Parallelized DNA Motif Search on
FPGA .Bioinformatics – Trends and
Methodologies.INTECH.
Faheem, H. M., 2010. “Associative Memory Array
Processor for Solving Motif Finding Problem”. The
International Conference on Artificial Intelligence and
Applications (AIA). Austria.
Rajasekaran, S., Balla, S. and Huang, C.H., 2005.Exact
algorithm for planted motif challenge problems.
Proceedings of Asia-Pacific Bioinformatics
Conference, 249–259.
Shunmaganathan, K., Deepika, K., Deeba, K., 2008.
Agent Based Bioinformatics Integration using
RESTINA. The International Arab Journal of
Information Technology. 5(3):258-264.
Konwinski, A., 2012.Multi-agent Cluster Scheduling for
Scalability and Flexibility. Technical Report
No.UCB/EECS-2012-273.
Russel, S., Norvig, P. 1995. Artificial Intellegence – A
Modern Approach.Printice-Hall.
Inta, R., Bowman, D., and Scott, M., 2012. The
“Chimera”: An Off-The-Shelf CPU / GPGPU / FPGA
Hybrid Computing Platform. International Journal of
Reconfigurable Computing. Vol. 2012.
AMultiagent-basedFrameworkforSolvingComputationallyIntensiveProblemsonHeterogeneousArchitectures-
BioinformaticsAlgorithmsasaCaseStudy
533
