
Essential Proteins and Functional Modules in the Host-Pathogen
Interactions from Innate to Adaptive Immunity
C. albicans-zebrafish Infection Model
Chia-Chou Wu and Bor-Sen Chen
Control and Systems Biology Laboratory, Department of Electrical Engineering,
National Tsing Hua University, Hsinchu, Taiwan
Keywords:
Computational Systems Biology, Network construction, Host-pathogen interaction, Protein-protein interac-
tion network, Infection.
Abstract:
The host and the pathogen are indispensable in the infectious diseases. Besides studying the host defensive and
pathogen invasive mechanisms individually, the cross-species interactions, i.e., the host-pathogen interactions,
become a novel and intense research subjects of the infectious diseases. In this study, two host-pathogen inter-
action networks are constructed for innate and adaptive immunity based on the time course microarray data of
C. albicans-zebrafish infection model. The interaction variations in the host, pathogen, and host-pathogen re-
gions are evaluated by comparing the two constructed networks. Those proteins of larger interaction variations
stand for more pivotal roles in the transition from innate to adaptive immunity. Moreover, in the host-pathogen
region, four significantly enriched functional modules are identified. Meanwhile, the interaction variations of
these four functional groups imply the corresponding strategy shifts of the host and pathogen from innate to
adaptive immunity. In view of these results, this study gives a systematic explanation about the transition from
innate and adaptive immunity from functional modules perspective. Thus, this study provides potential targets
for developing efficient therapies of the infectious diseases.
1 INTRODUCTION
The host and the pathogen are indispensable in the in-
fectious diseases. In particular,the interplays between
the host and pathogen shape the whole infection pro-
cesses from the first pathogens exposure to the final
outcomes of the infection (Tierney et al., 2012). After
activating the first line of host defense mechanisms,
the innate immunity recruits several types of cells
(e.g., macrophages, dendritic cells, NK cells, etc.)
to protect the host from the pathogen invasion and
then striveto eliminate the threats from the pathogens.
In turn, pathogens have evoked multiple strategies
for surviving under the host immune-defense mech-
anisms. After several rounds of attacks and defenses
between the host and pathogen, the host may elimi-
nate pathogens and prepare for the next challenges or
the pathogens may win the battle to cause chronic in-
flammation or death of the host. Despite the tremen-
dous advances in the pathogenic mechanisms and the
following triumph in the drug development (Arnold
et al., 2012), the remaining issues (e.g., drug resis-
tance) of infectious diseases become more trouble-
some. The dynamic and complex interactions be-
tween the host and pathogen may partially explain
why those drugs are often not effective in vivo (Mei-
jer and Spaink, 2011). Until a decade ago, the tradi-
tional viewpoint to treat the host and pathogen sepa-
rately is shifted to a more holistic viewpoint on both
players in the infection processes. This viewpoint
transition results from (i) the realization of the indis-
pensableness of the host-pathogen interactions (HPIs)
in the infectious diseases and (ii) the advent of the
OMICs biotechnology in measuring the genes, tran-
scripts, and proteins at whole cell/organism levels
(Schmidt and Volker, 2011). This permits a compre-
hensive interrogation of the pathogen at the whole-
genome, transcriptome, and proteome levels as well
as the host. From the molecular aspect, the infec-
tion processes can be viewed as the interference of
pathogenic proteins with the hosts’ interaction net-
work (Arnold et al., 2012). Hence, to investigate in-
fection processes from a systematic perspective, in
this study we would construct dynamic host-pathogen
protein-protein interaction (PPI) networks.
Regarding the success of the C. albicans-zebrafish
infection model (Chao et al., 2010) as well as the
17
Wu C. and Chen B..
Essential Proteins and Functional Modules in the Host-Pathogen Interactions from Innate to Adaptive Immunity - C. albicans-zebrafish Infection Model.
DOI: 10.5220/0005201700170025
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2015), pages 17-25
ISBN: 978-989-758-070-3
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

amenability to genetic manipulations (Gratacap and
Wheeler, 2014), the zebrafish is a novel and poten-
tial model organism to study the immunity. Fur-
thermore, the zebrafish and human immune systems
are remarkably similar and more than 75% of hu-
man genes implicated in diseases have counterparts
in zebrafish (Schier, 2013). This provides a strong
connection between the zebrafish and human on the
pathogenic mechanisms as well as immune responses,
which are important for biomedical applications. The
immune system of zebrafish as well as other verte-
brates can further be divided into two subsystems,
i.e., innate (unspecific) and adaptive (specific) im-
munity (Trede et al., 2004). Hence, the first dataset
we used to construct the dynamic host-pathogen PPI
network (HP-PPIN) measured the gene expression
profiles during the first 18 hours after zebrafish is
firstly exposed to a lethal dose of C. albicans (Chen
et al., 2013). This dataset sampled the gene ex-
pression profiles at 9 time points (i.e., 0.5, 1, 2, 4,
8, 12, 16, 18-hour post-injection) with three repli-
cates for C. albicans and zebrafish, respectively (Fig-
ure 1A). The outcomes of the interplay between the
host and pathogen after their first contact are cap-
tured by time course microarray experiments. Im-
mediately after the injection of C. albicans into ze-
brafish, the immune surveillance system of zebrafish
senses the existence of the invaders. The recogni-
tion of the pathogen-associated molecular patterns
(PAMPs) and/or damage-associated molecular pat-
terns (DAMPs) by the pattern recognition receptors
(PRRs) (e.g., toll-like receptors, C-type lectin recep-
tors, etc.) (Trede et al., 2004; Romani, 2011) may be
viewed as a starting point of a series of complex HPIs.
Those PRRs would initiate downstream pathways that
promote the activation of other parts of innate im-
mune system and the clearance of pathogens (e.g.,
production and secretion of cytokines, chemokines,
and chemotactic cues to recruit more immunocytes).
Thus, the morphological transitions (yeast-to-hyphal
form) (Kuo et al., 2013), required ions and small
molecules transportation (Wang et al., 2014), and
structures and components of molecules on the cell
wall changes (Romani, 2011) are the strategies uti-
lized by C. albicans to acquire nutrients and evade the
clearance from the host innate defensive mechanisms.
Those primary responses of the host and pathogen are
recorded in the first dataset which is a result from a
constant innate immune response and a delayed adap-
tive immune response (see Figure 1).
In addition to the activation of innate defensive
mechanisms, the PRRs and antigen presenting cells
would further activate the specific cells to clear the
pathogens much more efficiently (Romani, 2011). To
investigate the host-pathogen interactions in the spe-
cific defensive mechanisms (i.e., the adaptive immu-
nity), we adopted the second dataset. The experimen-
tal design of the second dataset comprises two C. al-
bicans injections into zebrafish. In the first injection,
a nonlethal dose is used to cause the host primary re-
sponses and immunological memory in the host. And
in 14 days after the first injection, a lethal dose is
applied to zebrafish. Then, the gene expression pro-
files are recorded through microarray after the second
injection and comprise 8 time points (2, 6, 12, 18,
24, 30, 36, 42-hour post-reinjection) with two repli-
cates for C. albicans and zebrafish, respectively (Fig-
ure 1A). Due to the previous exposure to C. albicans,
the immunological memory in zebrafish would be ac-
tivated and force the activation of the adaptive im-
munity. Thus, the main portion of the information
recorded in the second dataset reflects the outcomes
of the HPIs in the adaptive immunity. In contrast
to the innate immunity, the adaptive immunity of ze-
brafish and the responses of C. albicans to the adap-
tive immunity are less well-known. Here, in this study
the combination of these two datasets provides an op-
portunity to investigate the HPIs and their roles in the
innate and adaptive immunity.
The infection processes are often described as bat-
tles between the host and pathogen (Leroy and Raoult,
2010). The simultaneous considerations on the roles
of the host and pathogen in the infection processes
and the available genome-wide measurements make
the possibility of the systematic viewpoints on the ef-
fects of the HPIs in the innate and adaptive immu-
nity (Wang et al., 2013). In this study, the usage of
the C. albicans-zebrafish infection model (Chao et al.,
2010) shed light on the infectious diseases of the hu-
man host. As for the pathogen, the C. albicans is
the most virulent member of the CUG clade of yeasts
and a common cause of both superficial and invasive
infections (Lohberger et al., 2014) which may cause
life-threatening infections in immune-comprised host
(Odds, 1979), such as HIV positive patients. Investi-
gating the infection processes of C. albicans in detail
can improve the knowledge of the pathogenic mecha-
nisms and promote the control of infectious diseases.
Hence, the two datasets (GSE32119 and GSE51603)
are used for further analyses on the HPIs. To extract
the interaction information from the time course mi-
croarray data, two dynamic HP-PPINs are built up
for innate and adaptive immunity (Wang et al., 2013).
The HP-PPINs consist of the PPIs between zebrafish
and C. albicans, zebrafish and zebrafish, and C. albi-
cans and C. albicans. Through examining the interac-
tion variation between the innate and adaptive PPINs,
the interactions of the largest difference in the net-
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
18

work indicates the occurrences of the most dramatic
change. By evaluating the average interaction vari-
ation per edge, the critical proteins of the high in-
teraction variations in the interface of the host and
pathogen can be identified. Moreover, taking the ad-
vantage of advances in the ontology analysis, the sig-
nificantly enriched functional modules in the interface
of the host and pathogen can also be identified. Those
functional modules may imply the strategies taken by
the host and pathogen in the battles, the infection pro-
cesses. Thus, those function modules and the pro-
teins are potential drug targets of infectious diseases
(Schmidt and Volker, 2011).
2 MATERIALS AND METHODS
2.1 Overview of Microarray Data
In this study, we adapted two microarray datasets:
one is the temporal gene expression profiles of the
host (zebrafish) and pathogen (C. albicans) in the pe-
riod that they are firstly exposed to each other; the
other is the temporal expression profiles of the host
and pathogen in the period that they are secondly ex-
posed to each other. In the first set of microarray data,
the microarray experiments were performed to simul-
taneously profile genome-wide gene expressions in
both C. albicans and zebrafish during the infection
processes. Adult AB strain zebrafish were intraperi-
toneally injected with 1 × 10
8
C. albicans (SC5314
strain) cells (a lethal dosage). Whereas the second
microarray data measured the genome-wide gene ex-
pression level of the host and pathogen since their
second contact (with 1 × 10
7
C. albicans injection,
also a lethal dosage), that is, fourteen days after their
first contact (with 1 × 10
5
C. albicans, a non-lethal
dosage). Then, a two-step homogenization/mRNA
extraction procedure was performed using the whole
zebrafish infected with C. albicans. This approach
could provide separate pools of gene transcripts from
both the host and the pathogen, enabling individual
estimation of specific gene expression profiles in ei-
ther the host or the pathogen using sequence-targeted
probes derived from the individual genome. Agi-
lent in situ oligonucleotide microarrays, which cover
6,202 and 26,206 genes for C. albicans and zebrafish
respectively, were used to profile time-course gene
expression at 9 time-points (0.5, 1, 2, 4, 6, 8, 12, 16,
18 hours post-infection) with three replicates for both
organisms in the first microarray dataset (Chen et al.,
2013) and 8 time points (2, 6, 12, 18, 24, 30, 36, and
42 hours post-second infection) with two replicates
for both organisms in the second microarray dataset.
The first set of microarray was downloaded from the
GEO database (GSE32119) and the second set of mi-
croarray (GSE51603) was prepared under the similar
condition as the first set. Manipulation of the animal
model was approved by the Institutional Animal Care
and Use Committee of National Tsing Hua University
(IRB Approval No. 09808).
2.2 Protein Pool Selection and Database
Integration
There are two things to be completed before con-
structing the dynamic protein-protein interaction
(PPI) network. The first is to have a protein pool from
which the nodes in the resultant networks are cho-
sen. And the second step is to have all possible PPIs
among the proteins in the protein pool through in-
tegrating the interaction information from databases.
Here, our protein pool is consisted of the union of
the differentially expressed genes in the first and sec-
ond set of microarray data and the differentially ex-
pressed genes between the first and second microar-
ray datasets. The criterion to select the differentially
expressed genes in the first and second microarray
datasets is to compute the p-value of ANOVA test
whether the average expression levels are different
along the time (i.e., for the first dataset, the null hy-
pothesis is µ
1
= · ·· = µ
9
and for the second dataset,
the null hypothesis is µ
1
= · ·· = µ
8
) and then to se-
lect those proteins with the corrected p-value<0.05
into the protein pool. Also the genes in the top 5% of
the expression difference between the first and second
datasets were chosen into the protein pool. Next, for
the all possible interactions among the proteins in the
protein pool, the interaction information of zebrafish-
zebrafish, C. albicans-C. albicans, and zebrafish-C.
albicans are needed. However, the lack of the infor-
mation about these three kinds of interaction infor-
mation makes it difficult to collect all possible inter-
actions. Also it is impossible to consider full inter-
actions among the proteins in the protein pool. To
overcome the issue, the interaction information from
the human and yeast are used due to their similarity
to our studying subjects (zebrafish and C. albicans)
and data availability. To infer the possible interac-
tions of the studying subjects (zebrafish and C. albi-
cans), the orthologs information in the Inparanoid is
used to convert the interactions of human and yeast
into the interactions of zebrafish and C. albicans. It
should be noticed that the interactions inferred from
the ortholog-based method were derived under many
different experimental conditions, which cannot accu-
rately reflect the actual condition of host-pathogen in-
teractions during C. albicans infection processes; that
EssentialProteinsandFunctionalModulesintheHost-PathogenInteractionsfromInnatetoAdaptiveImmunity-C.
albicans-zebrafishInfectionModel
19

is, there exist false positives interactions in the all in-
ferred possible interactions of zebrafish and C. albi-
cans.
2.3 Host-Pathogen Protein-Protein
Interaction Network (HP-PPIN)
Construction
To construct the interspecies network from the protein
pool and inferred interactions, the dynamic model of
the protein-protein interaction is used to determine the
realistic interaction network with one protein by one
protein fashion. For a target protein i of the host, the
dynamic interaction model is as follows (Wang et al.,
2013):
p
(h)
i
[k+ 1] =σ
(h)
i
p
(h)
i
[k] +
N
∑
n=1
α
(h)
in
p
(h)
n
[k]
+
M
∑
m=1
γ
im
p
(p)
m
[k] + β
i
+ ε
i
[k+ 1]
(1)
and of the pathogen
p
(p)
i
[k+ 1] =σ
(p)
i
p
(p)
i
[k] +
M
∑
m=1
α
(p)
im
p
(p)
m
[k]
+
N
∑
n=1
γ
in
p
(h)
n
[k] + β
i
+ ε
i
[k+ 1]
(2)
where p
i
[k] denotes the protein activity level at time
k, the superscript of p
i
indicates the species of pro-
teins (h: host, zebrafish; p: pathogen, C.albicans),
ε
i
[k] denotes the environment noise at time k, σ
i
de-
notes the self-regulation ability, α
in
denotes the regu-
lation ability from the regulator n of the same species
as the target protein to the target protein i, γ
im
denotes
the regulation ability from the regulator m of the other
species to the target protein i, and β
i
denotes the basal
activity level of target protein i. The biological mean-
ing of this formulation is the protein activity level of
target protein i in the future (at time k + 1) is deter-
mined by the current protein activity level (at time k)
of itself (σ
i
), the regulations from other proteins of
the same species (α
in
) and the other species (γ
im
), the
basal activity, and the environmental noise. Due to
the unavailability of the proteomic data, the expres-
sion levels measured by the microarray experiments
are used to stands for the activity levels in the formu-
lation. The dynamic model for the host target protein
i can be further rewritten into a concise form as fol-
lows:
p
(h)
i
= Φ
i
θ
i
+ ε (3)
where
p
(h)
i
=
h
p
(h)
i
[1] ··· p
(h)
i
[K + 1]
i
T
,
θ
i
=
α
i1
··· α
iN
γ
i1
··· γ
iM
σ
i
β
i
T
,
ε
i
=
ε
i
[1] · ·· ε
i
[K + 1]
T
,
and
Φ
i
=
p
(h)
i1
[0] ··· p
(h)
iN
[0] p
(p)
i1
[0] ··· p
(p)
iM
[0] p
i
[0] 1
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
p
(h)
i1
[K] · ·· p
(h)
iN
[K] p
(p)
i1
[K] · ·· p
(p)
iM
[K] p
i
[K] 1
Similarly, the dynamic model for the pathogen can
also be rewritten into a similar form. The only un-
known parameter θ
i
can then be estimated by param-
eter estimation methods, such as least square estima-
tion. However, due to the lack of large scale measure-
ment of host and pathogen proteins, we alternatively
used gene expression profiles as a substitute of pro-
tein activities to identify the parameters in the model.
Furthermore, to make sure the model is unnecessar-
ily complex, the Akaike information criterion (AIC)
is introduced for model selection to balance the com-
peting objectives of conformity to the data and par-
simony, i.e., a trade-off between the model error and
model complexity. Hence, the final network encom-
pass the dynamic models of each protein with the
minimum AIC values.
2.4 Relevance Score Calculation
To target the essential proteins in the host-pathogen
protein-protein interaction network, the relevance
scores are calculated for proteins or functional mod-
ules to correlate proteins with the evolution of the
host-pathogen interactions from innate to adaptive
immunity. The relevance score is basically a measure-
ment of the variation of the regulation activity under a
condition transition. According to the dynamic mod-
els, the constructed PPI network under a specific mi-
croarray experiment condition can be written as fol-
lows:
p
(h)
1
[k+ 1]
.
.
.
p
(h)
N
[k+ 1]
p
(p)
1
[k+ 1]
.
.
.
p
(p)
M
[k+ 1]
=
σ
(h)
1
··· α
(h)
1N
γ
11
··· γ
1M
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
α
(h)
N1
··· σ
(h)
N
γ
N1
··· γ
NM
γ
11
··· γ
1N
σ
(p)
1
··· α
(p)
1M
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
γ
M1
··· γ
MN
α
(p)
M1
··· σ
(p)
M
×
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
20
p
(h)
1
[k]
.
.
.
p
(h)
N
[k]
p
(p)
1
[k]
.
.
.
p
(p)
M
[k]
+
β
(h)
1
.
.
.
β
(p)
1
.
.
.
β
(p)
M
+
ε
(h)
1
[k+ 1]
.
.
.
ε
(h)
N
[k+ 1]
ε
(p)
1
[k+ 1]
.
.
.
ε
(p)
M
[k+ 1]
(4)
or in a more concise form:
p[k+ 1] = Ap[k] + β + ε[k+ 1] (5)
where A is a matrix representation of the network con-
structed under a specific microarray experiment con-
dition. The regulation ability difference of two PPI
networks between innate and adaptive immunity can
be expressed as the following interaction difference
matrix form (Wang et al., 2014):
D
cond2−cond1
= A
cond2
− A
cond1
(6)
In the condition transition, if the variation of the reg-
ulation abilities of a protein is larger, it may implies
the protein plays a more important role in the condi-
tion transitions. So the relevance score of a protein
can be defined as follows:
RS
p
=
∑
Q
q=1
kd
pq
k
Degree of protein p
(7)
where d
pq
is the pq-entry of D
cond2−cond1
, that is, the
average regulation ability variation of the protein p.
The degree of protein p is the number of non-zero ele-
ment in pth row of the difference matrix D
cond2−cond1
.
The relevance score is proposed to evaluate the inter-
action variations of proteins in the network.
3 RESULTS
3.1 Overview of the Host-Pathogen
Protein-Protein Interaction
Networks (HP-PPINs) for Innate
and Adaptive Immunity
In this study, we aimed to understand the roles of
host-pathogen interactions in the innate and adaptive
immunity and the transition from innate to adaptive
immunity with a systems biology approach. The out-
comes of host-pathogen interactions are represented
in the simultaneous measurements of the temporal
gene expression profiles during the periods when in-
nate and adaptive immunity are activated (Figure 1A).
The HPIs in these two periods are then extracted by
Primary response
Day
0
14
Magnitude of response
Data 1
Data 2
Secondary response
(A)
Innate Adaptive
PathogenHost
0.5 1 2 4 6 8 12 16 18 2 6 12 18 24 30 36 42
hours post-reinjectionhours post-injection
(B)
(C)
10
8
CFU C. albicans
10
5
CFU C. albicans 10
8
CFU C. albicans
Temporal gene expression
profiles of innate immunity
(Data 1)
Inferred PPIs from databases
and ortholog information
Innate dynamic PPI
network
Adaptive dynamic PPI
network
Temporal gene expression
profiles of adaptive immunity
(Data 2)
Protein pool
Feature selection
Parameter identification
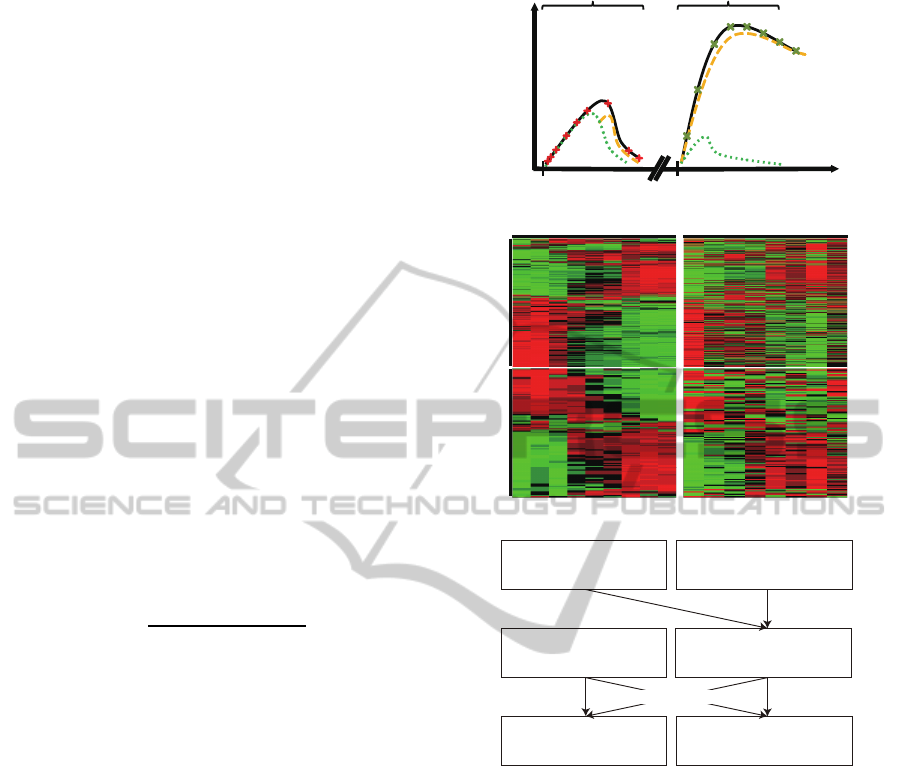
Figure 1: The overview of microarray experiments. (A) The
schematic of two microarray datasets. In the first dataset,
gene expression profiles of the C. albicans and zebrafish
are recorded at 9 time points (+). It mainly represent the
outcomes of HPIs in the innate immunity (green dot line).
In the second dataset, gene expression profiles of the C. al-
bicans and zebrafish are recorded at 8 time points (×). It
mainly represents the outcomes of HPIs in the adaptive im-
munity (yellow dash line). (B) The overview of microarray
data. (C) Flowchart for network construction. Features se-
lected from the two microarray datasets consist the protein
pool. The parameters in the dynamic models are identified
based on the expression profiles and the interaction infor-
mation of the selected features. In the end, two dynamic
HP-PPINs are constructed for innate and adaptive immu-
nity.
the dynamic models of the microarray data and fur-
ther visualized as two HP-PPINs (Figure 1B). The
protein pool encompassed 1620 proteins of interest
including differentially expressed features and the top
5% of the expression level difference between innate
and adaptive immunity (see Figure 1C) and there are
EssentialProteinsandFunctionalModulesintheHost-PathogenInteractionsfromInnatetoAdaptiveImmunity-C.
albicans-zebrafishInfectionModel
21

26060 PPI candidates. Then using dynamic network
construction (see Figure 1C and methods for details),
the resultant networks are consisted of 1512 proteins
(1431 for the C. albicans; 81 for zebrafish) and 5722
PPIs (5510 for the intracellular region of C. albicans;
145 for the interspecies interaction; 66 for intracellu-
lar region of zebrafish) for innate immunity and 1578
proteins (1480 for the C. albicans; 98 for zebrafish)
and 3755 PPIs (3577 for the intracellular region of C.
albicans; 96 for the interspecies interaction; 82 for
intracellular region of zebrafish) for adaptive immu-
nity. The details of the amount of nodes and edges
are summarized in the Figure 2B. In the amount vari-
ation of the nodes and edges of the pathogen, al-
though there exists plenty of nodes shared by both in-
nate and adaptive immunity in the host-pathogen and
pathogen-pathogen regions (Figure 2B), the number
of edges has changed from 5511 to 3577, that is, only
1203 edges are shared (Figure 2B). This implicates
that the pathogen may use the almost the same set of
protein (∼85%) but the different links to interact with
the host and regulate functions within the pathogen it-
self under different challenges at innate and adaptive
immunity. In contrast, the host might use a slightly
different strategy for self-regulation. To efficiently
identify and evaluate the importance of proteins in the
innate and adaptive immunity, we aggregated the two
networks (the innate and the adaptive immunity net-
work) into an interaction difference network (IDN),
i.e., the matrix D in the method (Figure 2A).
3.2 The Essential Proteins and
Functional Modules based on the
Relevance Scores
The relevance score stated in the previous section is
a quantity to represent the average interaction varia-
tions per links of a protein, that is, the ratio of the to-
tal interaction variation of a protein to the number of
links possessed by the protein. Hence, the relevance
score is considered to evaluate the extent of the inter-
action variations which may be critical in this study
for the observation of the amount of the nodes and
edges in the IDN (Figure 2A), the difference between
innate HP-PPIN and adaptive HP-PPIN. Comparing
to the other similar calculation (Wang et al., 2014),
our proposed relevance score is more proper to evalu-
ate the interaction variation of proteins in the network
since the consideration of degree of protein excludes
the proteins have many links of little variations. In
the following we would focus on the proteins of the
top ten in relevance scores at three regions, that is,
the host-host, host-pathogen, and pathogen-pathogen
region, respectively.
Figure 2: Result summary of dynamic HP-PPI network con-
struction. (A) The difference of the constructed innate and
adaptive networks (Node color: blue and red stand for ex-
clusive existence in the innate and adaptive immune re-
sponses respectively and purple stands for coexistence in
both immune responses. Edge color: blue and red stands
for the attenuated and enhanced interaction, respectively.).
(B) The number of the nodes and edges in the two dynamic
HP-PPINs.
3.2.1 The Host-host Region
In the region of host-host interaction (Figure 3), the
top ten proteins in the relevance scores showed their
close relationships with innate and adaptive immune
responses. Extracting the ten proteins and their first
neighbors from the IDN, there are five components
in the host-host region. The biggest one is consisted
of f2, LOC798231, LOC793315, ace2, gnai1, and
their first neighbors. Starting from gnat2, a host G-
protein, also one end of HPIs has connections with
chemokines-related proteins (cck-c5a and si:dkey-
269d20.3) and chemotaxis-related proteins (ENS-
DARP00000105159 and ENSDARP0000111107).
Then angiogenesis- and coagulation-related proteins
(agt, ace2, f2, and ENSDARP00000098661) are con-
nected to the chemokines-related proteins. In the fi-
nal part of the components, there are three more pro-
teins, serine proteinase inhibitor (serpinc1), proki-
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
22

Angiogenesis;
Coagulation
Chemokine
Complements
system
Figure 3: The ten proteins highest in relevance scores in the
host-host region and their first neighbors. The nodes with
shadow are the proteins of higher relevance scores (The
meaning of node color, edge color, and line style are the
same as Figure 2.).
neticin (ENSDARP00000109666), and suppressor of
IKBKE 1 (sike1). In this component, the roles of
angiogenesis and coagulation are manifest in the in-
nate and adaptive immunity. The second compo-
nent mainly consists of complements (c7b, c8g, c8a,
c8b, c9) and vitronectins (vtna and vtnb). Given
the well-known roles of complement system in im-
munity, the vitronectins got researchers’ attention in
the field of immunity recently (Gerold et al., 2008).
The cd36 and apolipoproteins form the third compo-
nent. CD36 plays a pivotal role in macrophage foam-
cell formation and atherogenesis, which is reduced by
apolipoproteins. Although the last two components
are less documented, the versican (vcanb) and tank
are reported their roles on the inflammation (Wight
et al., 2014).
3.2.2 The Pathogen-Pathogen Region
In the pathogen-pathogen region (Figure 4), the ten
proteins with the highest relevance scores and their
first neighbors form a single component. In this com-
ponent, the importance of redox status in the innate
and adaptive immune responses is emphasized again
(Wang et al., 2014). ERG1, CAL0005908, MET10,
and GCV3 are all related to the redox status of C. al-
bicans. Also CAL0005225,ERG1, and SDS24 are re-
sponsible to the expansion of C. albicans due to their
functions on the budding, filament growth, and cell
cycle, respectively. Especially, MET10 is also respon-
sible to the responses to the stress from the host and
environment. Another major function in this compo-
nent is the transferase activity. MET2 is a homoser-
ine acetyltransferase which can transform homoser-
ine, a toxin for C. albicans, to another compound.
ARG3 would facilitate the production of citrulline,
which can induce the pseudohyphal morphogenesis.
The morphology transformation of C. albicans has
been proven to be important in the pathogenesis of
C. albicans. In the end, the hydrolase, CAF16, exerts
SSD1
CAL0003938
CAL0001390
KAR2
CAL0004978
CAL0004601
NPL3
NAM7
CCR4
CAL0005908
CAL0004040
GDH2
CAL0002811
BUD14
SMT3
MHP1
ARP9
CAL0004008
CAL0000592
CLC1
HAT2
SDS24
ERG24
BNI1
ERG1
CAL0004558
CAL0004693
CDC7
ACT1
CAL0003932
SEC2
PGA4
GNP1
PHR2
CAL0000788
FUM11
GCV3
SCL1
ACH1
CAL0003509
GAP2
CAF0007087
ARG3
CAL0005816
RPN1
UAP1
RPN10
KSP1
CAL0005225
ZDS1
CDC28
CAF16
CLA4
MKC1
AAT21
ISW2
CAL0005376
CAL0004824
FGR32
PEX13
MFG1
RBK1
CYP5
CAL0002898
MET10
CAL0000831
CAL0003798
AKL1
OFD1
HAT1
MET2
PHO85
Redox status
Budding, filament growth, and cell cycle
Figure 4: The ten proteins highest in relevance scores in
the pathogen-pathogen region and their first neighbors (The
meaning of node color, edge color, and line style are the
same as Figure 2.).
its influence on the RNA polymerase II although the
specific affected genes are still unknown.
3.2.3 The Host-Pathogen Region
In the host-pathogen region, we also selected 10 pro-
teins from the host and the pathogen, respectively.
Those interspecies proteins form more complicated
interaction networks in the host-pathogenregion (Fig-
ure 5). Meanwhile, the functional structure is slightly
different comparing to the host-host and pathogen-
pathogen regions. A possible mechanism how redox
status in the host and pathogen correlates is shown
in the extracted IDN, i.e., the interaction between
thioredoxin (txn) and RiboNucleotide Reductase 1
(RNR1). In addition to the redox role of RNR1,
RNR1 also has impact on the iron utility, filament
growth, and cell cycle. This implicates the effect of
redox status on the pathogen is multifaceted. One
of the interactions of RNR1, the link between RNR1
and CAL0003932, is attenuated in the adaptive im-
mune response. This implies the ability of adap-
tive immunity to attenuate the deubiquitination and
degradation of proteasome in the pathogen. How-
ever, CAL0003932 has not been well-characterized.
A group of chemokine-related host proteins consti-
tutes another similar function in the host-pathogen re-
gion. Comparing to the chemokine-related function
in the host-host region, its roles in the HPIs are more
interesting. CAG1, the entry how chemokine-related
functions affect the pathogen, is related to the hyphal
growth, mating, and biofilm formation of pathogen,
which are all important in the pathogenesis. Besides
the same functions (redox status and chemokines) in
the pathogen-pathogen and host-host regions, there
are several functions which can only be seen in the
host-pathogen region, that is drug responses, fatty
acid, glycine metabolism, and circadian. The inter-
actions among TAF60, gtf2a2, polr2e emerge in the
adaptive immunity. TAF60, a transcription factor, is
EssentialProteinsandFunctionalModulesintheHost-PathogenInteractionsfromInnatetoAdaptiveImmunity-C.
albicans-zebrafishInfectionModel
23

RPM2
per1b
cry2b
AMS1
DOG1
per2
arntl1b
cry2a
pygb
gys2
azi1
CAR2
HRR25
DCP2
SES1
NIP1
CAL0001603
ERG20
IPL1
EMP24
CAL0003394
HEM15
FAS2
CAL0001016
CAF0006974
CAL0004978
mhc1uba
PDX3
LAT1
ORC1
SHM1
VMA4
OLE1
actr3b
CAL0006149
CAL0005659
FUM11
SEC24
NSP1
UAP1
CAL0002752
CAL0000821
DPS1-1
CDC60
FAA4
GCV1
CDC28
CAL0002889
shmt1
RPN10
SYS1
RIC1
RPN1
VPH1
CAL0002020
GLN3
CAL0003403
YME1
PHR2
CAL0001938
TRK1
CAL0002617
CAL0000581
RHO3
SNZ1
ARF2
CAL0003207
CAL0000524
MES1
ERG26
UBP13
CAL0004665
CAL0006275
VAC8
CAL0000165
polr2eb
GSC1
gtf2a2
ADE2
FUN31
CAL0001155
PAP1
CAG1
gnai1
gnat2
STE18
ENSDARP00000111107
ENSDARP00000105159
ccl-c5a
si:dkey-269d20.3
CAL0000531
CHO1
CAL0003208
CAL0003987
CAL0004670
CAL0002919
ADE13
CAL0000243
GNP1
ERG1
CAL0002715
HEM13
CAL0003071
TPK2
CAL0006120
BRE1
BCY1
ppp1r1b
fabp1b.1
CMP1
GCV2
ABP1
CMD1
CUE5
CAL0001326
TAF60
SCS7
TAF19
CAL0003530
JAB1
si:ch211-204c21.1
GLO3
CAF0007003
HIS1
GRE3
MET15
GLN4
PGK1
CAL0006081
RPT6
GPA2
ARO8
CAL0004190
CAL0000101
GDE1
SIN3
FBA1
CAL0004479
MNT1
TKL1
CAL0003932
ARG1
ARO3
CTA3
MNT2
CAL0003043
SEC18
CAL0001235
CAL0002259
RPN2
CAL0000804
CAL0005635
ARG5,6
HSP70
ILV5
CAL0003768
SPT5
CAL0004266
SSD1
GVP36
CAL0001316
CAL0003837
CAL0006116
GDI1
CAL0005180
LEU1
CAL0004574
CAL0003861
CAL0001798
eif5b
NAM7
ISN1
NPL3
SSU81
CAL0001588
CAL0005131
CAL0000927
MCM1
CAL0000380
CCT3
CAL0000827
dusp4
HEM14
CAF0007058
NTH1
CEK1
CAL0004008
txn
PKC1
CAL0000331
smurf1
CAL0004305
URA7
THR1
CRM1
LEU4
AKL1
ESA1
POL3
ENSDARP00000076901
RNR1
RAD52
SHM2
HSM3
CAL0004051
CAL0006158
CAF0007423
GLK1
ILV2
CEF3
MSH6
MET13
ARG3
SMT3
pold1
POL2
RAD32
RSP5
PMS1
Chemokines
Circadian
Figure 5: The ten host and pathogen proteins highest in rele-
vance scores in the pathogen-pathogen region and their first
neighbors (The meaning of node color, edge color, and line
style are the same as Figure 2.).
responsible for the drug responses in the pathogen and
gtf2a2 and polr2e are related to the gene transcription
in the host. Their interactions stand for a possible
mechanism how the effects of HPIs are exerted into
the gene level. Other proteins for drug response are
SEC24 and LEU4, which are not well-characterized.
The fatty acid binding protein (fabp1b.1) of the host
shows the involvement of fatty acid in the innate and
adaptive immune responses. This is another proof for
the hypothesis of a local and systemic crosstalk be-
tween adipocytes and monocytes mediated by fatty
acids (Kopp et al., 2009). This fatty acid binding
protein links to the protein of the pathogen, SIN3,
related to the filament growth. The interaction be-
tween fatty acid binding protein and SIN3 becomes
more negative in the adaptive immunity. The third
function, glycine metabolism, has been implicated the
contribution to the infectious capacity of the pathogen
(Flynn et al., 2010). The final and interesting function
is the circadian rhythm in the host and pathogen. The
circadian rhythm-related proteins of the host (cry2a,
cry2b, and per2) and the pathogen (HRR25) form a
sub-network in the host-pathogen region. The circa-
dian rhythms in the host and pathogen are correlated
and there are plenty of the pathogens functions (yeast-
hyphal switch, gene transcription, pathogenesis, etc.)
are affected through HRR25.
4 CONCLUSION
In this study, the dynamic network modeling is used
to identify the complex and dynamic HPIs during two
different types of immune system. Based on the high-
throughput expression level measurements, two HP-
PPINs are constructed and then compared with each
other. We found that the pathogen may change the in-
teractions between proteins rather than recruit a whole
new set of proteins to react with the host. Hence,
we proposed the relevance score to quantify the dif-
ference of the regulation of a protein between the
innate and adaptive HP-PPIN. The relationships be-
tween the HPIs and several proteins of higher rele-
vance scores are verified by literatures. Also, sev-
eral not well-known proteins but with higher rele-
vance scores are suggested their roles in the HPIs.
Moreover, the circadian-, redox status-, angiogenesis-
, and coagulation-related functions are correlated in
the HPIs. The interplays among the four functional
modules cause the changes in the mechanisms of the
pathogenesis, responses to stresses, and cell cycle in
the C. albicans. In the end, these proteins of higher
relevance scores and the four functional modules are
regarded as the essential components in the HPIs as
well as the potential targets for future drug develop-
ments.
ACKNOWLEDGEMENTS
Ministry of Science and Technology, R.O.C. (Taiwan)
[MOST 103-2745-E-007-001-ASP].
REFERENCES
Arnold, R., Boonen, K., Sun, M. G. F., and Kim, P. M.
(2012). Computational analysis of interactomes:
Current and future perspectives for bioinformatics
approaches to model the host-pathogen interaction
space. Methods, 57(4):508–518.
Chao, C. C., Hsu, P. C., Jen, C. F., Chen, I. H., Wang, C. H.,
Chan, H. C., Tsai, P. W., Tung, K. C., Wang, C. H.,
Lan, C. Y., and Chuang, Y. J. (2010). Zebrafish as a
model host for Candida albicans infection. Infection
and Immunity, 78(6):2512–2521.
Chen, Y. Y., Chao, C. C., Liu, F. C., Hsu, P. C., Chen, H. F.,
Peng, S. C., Chuang, Y. J., Lan, C. Y., Hsieh, W. P.,
and Wong, D. S. H. (2013). Dynamic transcript pro-
filing of Candida albicans infection in zebrafish: A
pathogen-host interaction study. PloS One, 8(9).
Flynn, N. E., Patyrak, M. E., Seely, J. B., and Wu, G. Y.
(2010). Glycine oxidation and conversion into amino
acids in Saccharomyces cerevisiae and Candida albi-
cans. Amino Acids, 39(2):605–608.
Gerold, G., Abu Ajaj, K., Bienert, M., Laws, H. J., Zychlin-
sky, A., and de Diego, J. L. (2008). A Toll-like recep-
tor 2-integrin β
3
complex senses bacterial lipopeptides
via vitronectin. Nature Immunology, 9(7):761–768.
Gratacap, R. L. and Wheeler, R. T. (2014). Utilization of
zebrafish for intravital study of eukaryotic pathogen-
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
24

host interactions. Developmental and Comparative
Immunology, 46(1):108–115.
Kopp, A., Gross, P., Falk, W., Bala, M., Weigert, J., Buech-
ler, C., Neumeier, M., Scholmerich, J., and Schaffler,
A. (2009). Fatty acids as metabolic mediators in in-
nate immunity. European Journal of Clinical Investi-
gation, 39(10):924–933.
Kuo, Z. Y., Chuang, Y. J., Chao, C. C., Liu, F. C., Lan,
C. Y., and Chen, B. S. (2013). Identification of
infection- and defense-related genes via a dynamic
host-pathogen interaction network using a Candida
albicans-zebrafish infection model. Journal of Innate
Immunity, 5(2):137–152.
Leroy, Q. and Raoult, D. (2010). Review of microarray
studies for host-intracellular pathogen interactions.
Journal of Microbiological Methods, 81(2):81–95.
Lohberger, A., Coste, A. T., and Sanglard, D. (2014). Dis-
tinct roles of Candida albicans drug resistance tran-
scription factors TAC1, MRR1, and UPC2 in viru-
lence. Eukaryotic Cell, 13(1):127–142.
Meijer, A. H. and Spaink, H. P. (2011). Host-pathogen in-
teractions made transparent with the zebrafish model.
Current Drug Targets, 12(7):1000–1017.
Odds, F. C. (1979). Candida and candidosis. University
Park Press, Baltimore.
Romani, L. (2011). Immunity to fungal infections. Nature
Reviews Immunology, 11(4):275–288.
Schier, A. F. (2013). Genomics zebrafish earns its stripes.
Nature, 496(7446):443–444.
Schmidt, F. and Volker, U. (2011). Proteome analysis of
host-pathogen interactions: Investigation of pathogen
responses to the host cell environment. Proteomics,
11(15):3203–3211.
Tierney, L., Kuchler, K., Rizzetto, L., and Cavalieri, D.
(2012). Systems biology of host-fungus interactions:
turning complexity into simplicity. Current Opinion
in Microbiology, 15(4):440–446.
Trede, N. S., Langenau, D. M., Traver, D., Look, A. T., and
Zon, L. I. (2004). The use of zebrafish to understand
immunity. Immunity, 20(4):367–379.
Wang, Y. C., Lin, C., Chuang, M. T., Hsieh, W. P., Lan,
C. Y., Chuang, Y. J., and Chen, B. S. (2013). Inter-
species protein-protein interaction network construc-
tion for characterization of host-pathogen interac-
tions: a Candida albicans-zebrafish interaction study.
BMC Systems Biology, 7.
Wang, Y. C., Tsai, I. C., Lin, C., Hsieh, W. P., Lan, C. Y.,
Chuang, Y. J., and Chen, B. S. (2014). Essential func-
tional modules for pathogenic and defensive mecha-
nisms in Candida albicans infections. Biomed Re-
search International.
Wight, T. N., Kang, I., and Merrilees, M. J. (2014). Versi-
can and the control of inflammation. Matrix Biology,
35:152–161.
EssentialProteinsandFunctionalModulesintheHost-PathogenInteractionsfromInnatetoAdaptiveImmunity-C.
albicans-zebrafishInfectionModel
25
