
An Enhanced DNA-based Steganography Technique
with a Higher Hiding Capacity
Samiha Marwan
1
, Ahmed Shawish
1,2
and Khaled Nagaty
1,2
1
Department of Computer Science, The British University in Egypt, Cairo, Egypt
2
Ain Shams University, Cairo, Egypt
Keywords:
Security, DNA-based Steganography, Data Hiding, Encryption, Decryption, Playfair Cipher.
Abstract:
DNA-based Steganography is one of the promising techniques to secure data exchange, where data is hidden
into a real DNA sequence. For the sake of security, some steganography techniques encrypt data before hiding
it which strengthen the technique’s steganalysis. One of the widely used encryption techniques is the DNA-
based playfair cipher. This technique intensively requires a long list of preprocessing steps in addition to
extra bits which must be added to guarantee successful decryption. Nevertheless, the succeeding hiding step
suffers from a limited capacity, which turns this current DNA-based Steganography technique into a complex,
inefficient, and time consuming process. In this paper, we propose a new DNA-based Steganography algorithm
to simplify the current technique as well as achieve higher hiding capacity. In the proposed algorithm, we
enhance the commonly used playfair cipher by defining a novel short sequence of preprocessing steps and
getting rid of the extra overhead bits. We also utilize a more efficient technique to enhance the hiding phase.
The proposed approach is not only simple and fast but also provides a significantly higher hiding capacity
with a high security. The conducted extensive experimental studies confirm the outstanding performance of
the proposed algorithm.
1 INTRODUCTION
In the world where information and communication
become indispensable, it becomes a must for research
to find out solutions for data protection, integrity and
accuracy. Recently, DNA-based Steganography be-
comes one of the promising techniques to secure data
exchange, where data is hidden into a real DNA se-
quence. The complexity of the DNA structure and
the randomness of its data are the main drivers of its
outperformance in comparison with other traditional
Steganography methods (Smith, 2003),(Alberts and
Johnson, 2008),(Adleman, 1994). For the sake of se-
curity, some DNA-based Steganography approaches
encrypt the data first through a ciphering technique
and then hide it into a real DNA sequence. This paper
focuses on such approach as it leads to a more con-
fusion to the attacker and strengthen the technique’s
steganalysis
1
.
The 5x5 playfair cipher is one of the well known
and commonly used substitution ciphering techniques
1
Study of identifying the existence of data and detecting
it.
that uses a 5x5 grid containing the English alphabet in
an ascending order from A to Z, where 24 letters oc-
cupies 24 cells and the remaining 2 letters -usually I
& J- occupy the remaining cell. The sender and the
receiver should agree on a specified keyword to rear-
range the ordering of its cells to guarantee the unique-
ness of the 5x5 grid each time the key is changed as
shown in Fig1. Recently, this ciphering technique is
used to encode DNA-based data due to its strong en-
crypting capability in comparison with the other en-
cryption techniques (Atito, A. et al., 2012). How-
ever, such technique come up with long list of pre-
processing steps that, in our point of view, do not
decrease the cracking probability but complicate the
implementation process and increase the processing
time. Nevertheless, it also decreases the hiding ca-
pacity as we will prove in the rest of this paper. All
these issues contradicts with the fact of the DNA’s
huge storage capacity.
In the current DNA-based 5x5 playair cipher
implementation (Khalifa and Atito, 2012), the target
message passes through a long sequence of transfor-
mations: from letters to binary, from binary to DNA
150
Marwan S., Shawish A. and Nagaty K..
An Enhanced DNA-based Steganography Technique with a Higher Hiding Capacity.
DOI: 10.5220/0005246501500157
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2015), pages 150-157
ISBN: 978-989-758-070-3
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

letters
2
, from DNA letters to protein sequence
presented by English letters
3
, where ciphering
technique takes place. Then the resulted ciphered
English letters are again transformed to DNA letters
with extra overhead bits, which is generally known as
the ambiguity bits. Finally, the resulted DNA letters
are concealed into a real DNA sequence through a
hiding technique. The whole process is then reversed
again at the receiver’s node to extract the original
message. As we can easily note, the whole process is
a set of complicated long steps that only consume a
lot of the computational effort without a real addvalue
to the security strength.
In this paper, we propose an enhanced DNA-based
Steganography algorithm that is much more efficient
and faster than the current technique with a higher
hiding capacity. In the proposed algorithm, we en-
hance the commonly used playfair cipher by defin-
ing a novel sequence of preprocessing steps and get-
ting rid of the overhead. We also utilize a more effi-
cient technique to enhance the hiding process (Khalifa
and Atito, 2012). The proposed algorithm has rede-
fined the whole process in a much smarter and straight
forward mechanism resulting in a better performance
and low execution time with a higher hiding capacity.
Moreover, The security strength has been carefully
checked and proved through the calculation of the
cracking probability. The outstanding performance of
our proposed algorithm is demonstrated through ex-
tensive experimental studies.
The rest of this paper is organized as follows. Sec-
tion 2 overviews the background and related work
on the current Steganography techniques. Section 3
presents the proposed technique in detail. Section 4
discusses its performance analysis. Finally, the paper
is concluded in Section 5.
2 BACKGROUND
In this section, we provide a brief review on the DNA
and the related work. In addition, we discussed in
detail the main problems of the current DNA-based
Steganography techniques and their problems.
2.1 DNA Overview
DNA is the magic code for life (Smith, 2003), it con-
tains the genetic instructions used in the development
and functioning of all living organisms. Inspired from
2
The DNA letters are A, G, C, and T.
3
The protein sequence is composed of amino acids, each
is abbreviated by an English letter.
Key = “PLAY”
A B C D E
F G H I/J K
L M N O P
Q R S T U
V W X Y Z
P L A Y B
C D E F G
H I/J K M N
O Q R S T
U V Q X Z
Figure 1: 5x5 Playfair Cipher Grid before and after using
the Key.
nature, the fact that DNA molecule carries all the ge-
netic information, evolves the idea of using DNA it-
self as a data carrier. The information in DNA is
stored as a code made up of four chemical bases
named as nucleotides: adenine (A), guanine (G), cy-
tosine (C), and thymine (T). The sequence of these
four bases encodes the genetic information (Alberts
and Johnson, 2008). Each of the three nucleotides is
called a codon, therefore in nature there are 64 codons
since there are (4x4x4) letter combinations.
DNA has two main advantages that make it effi-
cient for data hiding and transmission. First of all its
high storage capacity; as proved by(Adleman, 1994).
Secondly, the simplicity of converting data to DNA
sequence makes it a good choice for data encryp-
tion within it. By exploiting the advantages of a
DNA as an efficient data carrier in addition to using
a well-suited encryption technique, researches ended
up by many solutions for secure data communica-
tion and transmission. DNA steganography is one of
these promising solutions(Peterson, 2001), (Catherine
et al., 1999),(Leier et al., 2000),(Shimanovsky et al.,
2002),(SAEB et al., 2007).
2.2 Related Work
In 1999, (Catherine et al., 1999) started DNA
steganography, where data is encrypted in DNA and
hid into microdots. In 2000 (Leier et al., 2000) pro-
posed a hiding technique where data can be encoded
into DNA sequence, however the original data can be
easily recovered once the primer sequence is known.
In 2001 (Peterson, 2001) proposed another new
scheme for secret data hiding but unfortunately it
had some concerns as it can be cracked through a
frequency-based cryptanalysis technique. In 2010
(Shiu et al., 2010) proposed three reversible data hid-
ing schemes based on DNA sequence, the most signif-
icant one was the substitution method, yet its hiding
capacity is not efficient enough.
In May 2012 (Khalifa and Atito, 2012) proposed
a Steganography technique, where data is encrypted
using DNA-based playfair cipher, then hid in a real
DNA sequence using a modified substitution tech-
nique to increase its hiding capacity. Although it
achieved higher hiding capacity than the original sub-
AnEnhancedDNA-basedSteganographyTechniquewithaHigherHidingCapacity
151

Table 1: Example of mapping codons to characters.
Character Codons
F GCT, GCC, GCA
B TAA, TGA, TAG
C TGT, TGC
N AAT, AAC
P CCT, CCC,CCG
O TTA, TTG
R CGT, CGG,CGA, CGC
M ATG
stitution method, yet this hiding capacity was not ef-
ficient enough as a result of the ambiguity problem.
In October of the same year, (Taur et al., 2012)
proposed another modified substitution technique
achieving a high hiding capacity but without encrypt-
ing data which minimized its security.
2.3 Ambiguity Problem
One of the most critical cons of the current technique
is the ambiguity problem. Hereby, we are providing a
brief description of such problem.
In nature, each codon in a DNA sequence is con-
verted to one of the 20 amino acids forming a pro-
tein sequence that is responsible for a certain func-
tionality. Each amino acid is abbreviated by an En-
glish letter, i.e. ”Alanine” is an amino acid abbrevi-
ated with letter ”A”. Since We have 64 codons and
20 amino acids, each amino acid maps to at most
4 codons leading to the ambiguity problem. (Sabry
et al., 2010) solve this problem by adding two extra
bits next to each amino acid identifying which codon
it represents. (Table 1) is an example showing the
mapping of 8 characters to codons. For clarification
assume that we have DNA sequence composed of two
codons: ” GCC AAT”. This sequence when converted
to characters using (Table 1), it will be: ” F N ” .
In the decryption process when converting from
characters to DNA sequence, we will not know which
codon does character ”F” represent. This is solved by
adding 2 extra bits that represent a DNA base as clar-
ified in (Table 2), to identify which codon does the
character represent. Assume that base ”A” represents
the 1st codon, ”G” for the 2nd codon, ”C” for the 3rd
and ”T” for the fourth codon. so instead of having ”F
N” we will have ”FG NA” , where ”G” is an ambigu-
ity base refers to second codon and ”A” refers to first
codon.
2.4 Hiding using 5x5 Playfair Cipher
Technique
In this subsection we explain the currently used en-
cryption and decryption process using DNA-based
5x5 playfair cipher as well as the recent substitution
process used for DNA hiding mentioned in (Khalifa
and Atito, 2012).
2.4.1 Encryption and Decryption using the
Current DNA-based 5x5 Playfair Cipher
Technique :
The encrypting process works as follows:
1. Convert message text to the binary form where
each character is presented by 8 bits.
2. Transform the binary form into DNA letters using
(Table 2).
3. The DNA form is transferred to the Amino acids
letters representation according to the new al-
phabet distribution with the corresponding new
codons used in (Sabry et al., 2010), taking into
consideration two ambiguity bits for each Amino
acid letter.
4. Separate the ambiguity bits from the amino acids
sequence.
5. Use the key of Upper case letters to generate the
5X5 grid.
6. Apply the traditional Playfair cipher process.
7. Encrypted amino acids letters are transferred back
to DNA sequence form.
8. Concatenate the DNA sequence with the saved
ambiguity bits. Eventually we got an encrypted
DNA sequence.
The decryption process:
Given the key and the encrypted DNA sequence
1. Separate the ambiguity bits from the encrypted
DNA sequence.
2. Convert encrypted DNA sequence to amino acids
letters.
3. Use the key to generate the 5x5 playfair cipher
grid.
4. Perform the inverse of the playfair cipher process.
5. Use the ambiguity bits to get the correct DNA se-
quence.
6. Convert DNA sequence to binary form.
7. Convert the binary form to the original plaintext.
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
152

Table 2: DNA letter representation of binary bits.
DNA letter Binary representation
A 00
G 01
C 10
T 11
Although this 5x5 DNA-based playfair cipher
technique encodes encrypted data efficiently it suffers
from two drawbacks. First is the ambiguity problem,
which means that for every codon we are enforced to
add 2 more bits to solve the ambiguity problem i.e.
3/4 of the ciphered DNA is real data while 1/4 is
for solving the ambiguity problem, which minimizes
the hiding capacity. Second, the long list of unneces-
sary complicated iterations consume a lot of compu-
tational resources especially when dealing with large
data sizes.
2.4.2 Implemented Substitution Process
The above mentioned encryption technique uses
a modified substitution method to achieve the
Steganography goal. This technique is mentioned in
detail in (Khalifa and Atito, 2012), it proved to be
better than the original one mentioned in (Shiu et al.,
2010). It assumes that the length of cover DNA se-
quence is the same as the message itself (S), and ac-
cording to the 5x5 playfair cipher explained above,
only 3/4 of these bases represent the actual message
bits, since the remaining 1/4 are reserved for the am-
biguity bits. The hiding capacity is measured in terms
of the number of hidden bits per neuclotide
4
(bpn).
Since each DNA base actually represents two bits of
the binary message (M), therefore the hiding capacity
is represented by the following equation:
Capacity =
Sizeo f messageinbits
Sizeo f coverinbases
(1)
=
3
4
∗ |S| ∗ 2
|S|
=
3
2
bpn (2)
From the previous equation we got that the hid-
ing capacity of the current hiding technique using the
DNA-based 5x5 playfair cipher is 3 bits per 2 neu-
clotides.
3 PROPOSED TECHNIQUE
The proposed DNA-based Steganography technique
consists of two phases, First the ciphering phase,
4
the nucleoide is the same as the DNA-base
•
Construction of 4x4
PFC grid using Key
Msg to binary
conversion
Binary to English
letters conversion
Apply playfair cipher
process on English
letters
English letters to
DNA conversion
Encrypted Data
encoded in a DNA
sequence (Enc)
Construction of 4x4
PFC grid using Key
Binary to original
message
Enc to English letters
conversion
Apply the inverse of
playfair cipher process
on English letters
English letters to
Binary conversion
Encrypted Data
encoded in a DNA
sequence (Enc), Key
Figure 2: Overall Ciphering and Deciphering processes of
the proposed technique.
Table 3: Example of 16 randomly generated English letters
-Playfair cipher grid-.
H C M U
D G Z B
I A X J
Q V W F
where we used 4x4 playfair cipher grid as a mod-
ified version to the current 5x5 playfair cipher grid
(Sabry et al., 2010), (Khalifa and Atito, 2012), (Atito,
A. et al., 2012) to avoid all its drawbacks, Figure 2
shows a short, illustrative diagram to the overall pro-
posed ciphering and deciphering processes. Second,
the hiding phase using the modified substitution pro-
cess in (Khalifa and Atito, 2012). As a result of mod-
ifying the ciphering process we succeeded in achiev-
ing higher hiding capacity than (Khalifa and Atito,
2012). In the following subsections we discuss in de-
tail the proposed Steganography algorithm from both
the sender and receiver side, followed by a step-by-
step illustrative example.
3.1 Ciphering
In our proposed ciphering algorithm we used 4x4
playfair cipher grid. Since the English alphabet con-
sists of 26 letters, we will use the Key’s ASCII value
as a seed number for generating 16 unique random
English letters to be represented by the 4x4 playfair
Table 4: 4x4 Binary grid.
0000 0001 0010 0011
0100 0101 0110 0111
1000 1001 1010 1011
1100 1101 1110 1111
AnEnhancedDNA-basedSteganographyTechniquewithaHigherHidingCapacity
153
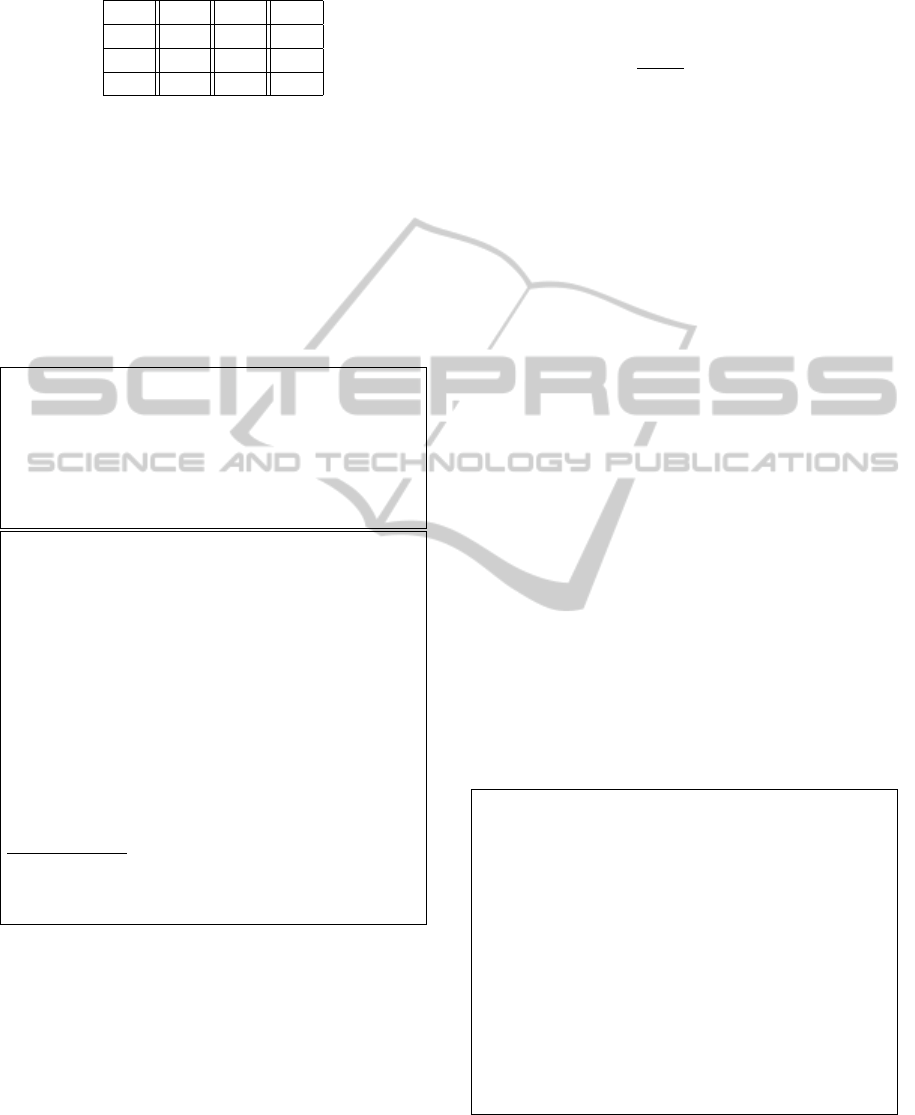
Table 5: 4x4 DNA grid.
AA TC CG TG
GC TT TA GT
GG AT CT CC
CA AC AG GA
cipher grid as the example shown in (Table 3).
Another 4x4 grid will be used, called the 4x4 binary
grid where each cell contains one of the 4-bit possible
combinations as shown in (Table 4), where the values
in this grid are ordered in an ascending manner.
Again we will construct another 4x4 grid, called the
4x4 DNA grid that contains the 16 possible 2-DNA
letters combinations, the initial order of these com-
binations is shown in (Table 5). Then the encryption
process done by the sender is implemented as follows:
Preprocessing (Key)
1. Use the Key as a seed value for generating 16
random English letters to form the 4x4 playfair
cipher grid.
2. Use the Key to shuffle the 4x4 binary grid and
the 4x4 DNA grid.
Encryption (Msg, 4x4-pfc-grid, ShuffledBG, Shuf-
fledDG)
a
1. Convert Msg to its binary form B.
2. Transform B to English letters E by mapping
each value in ShuffledBG to its corresponding
position in 4x4-pfc-grid.
3. Perform playfair cipher technique on E to get a
ciphered text C.
4. Map positions of C in 4x4-pfc-grid to its cor-
responding DNA letters in ShuffledDG lying in
the same cell position, to get the final encrypted
DNA sequence Enc.
a
pfc-grid stands for playfair cipher grid, ShuffledBG is
the shuffled Binary Grid, and ShuffledDG is the shuffled
DNA grid.
Note that, the values in the 4x4 binary grid and the
4x4 DNA grid are initial values which must be shared
between the sender and the receiver. The sender will
use the Key to shuffle these grids, where the resultant
shuffled grids will be used for encrypting the binary
data into DNA-based encrypted data.
3.2 Hiding Phase
After encryption, we applied the recent substitution
method (Khalifa and Atito, 2012) for hiding data to
achieve the goal of Steganography. By using equa-
tion(1) with the proposed encryption technique, we
will achieve higher hiding capacity as proved by the
following equation:
=
|S| ∗ 2
|S|
= 2bpn (3)
From the previous equation, we can see the great im-
pact of our proposed encryption technique on the sub-
stitution method. It has a significant higher hiding ca-
pacity, as a result of the ambiguity problem removal.
In other words, the hiding capacity is improved by
25% since in (Khalifa and Atito, 2012) it was 3/2 bits
per nucleotide, while in our proposed technique it is 2
bits per nucleotide.
3.3 Extraction
The extraction process is formed by the receiver, to
extract the hidden encrypted DNA sequence. In our
technique we used the extraction process in (Khalifa
and Atito, 2012).
3.4 Deciphering
The receiver will receive the encrypted DNA se-
quence and the Key through a secure channel. Then
s/he will use the Key to be able to shuffle the 4x4
binary grid and the 4x4 DNA grid to decrypt the
extracted encrypted DNA sequence. Ultimately, same
key must be used by the sender and the receiver. The
following is the proposed decryption algorithm which
is the reverse of the aforementioned Encryption algo-
rithm. It will be used by the receiver to retrieve the
original data from the encrypted DNA sequence.
Decryption(Enc, Key)
1. Perform the Preprocessing function mentioned
before the encryption step to obtain the 4x4-pfc-
grid, ShuffledBG and the ShuffledDG.
2. Map the positions of each 2 letters of the en-
crypted DNA sequence Enc in ShuffledDG to its
corresponding positions in 4x4-pfc-grid to get
English text C.
3. Perform the inverse of playfair cipher technique
on C to get E.
4. Map E to ShuffledBG to get its binary form B.
5. Convert B to the original message Msg.
Our proposed algorithm allows the message and
the key to be of any type. The used 4x4 matrix elimi-
nates the ambiguity problem that was presented in the
previous algorithms(Sabry et al., 2010), (Khalifa and
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
154

Table 6: 4x4 Shuffled Binary grid.
0111 0010 1100 0011
0110 0001 1000 0000
1001 0101 1010 1110
0100 1111 1011 1101
Atito, 2012). Additionally, it provides more simplic-
ity with no redundancy in the processes, which leads
to higher remarkable performance and lower execu-
tion time.
Moreover, as mentioned in the steps of our algo-
rithm, we use the numeral value of the key to gen-
erate random English letters to construct the playfair
cipher grid, which makes the playfair cipher tech-
nique more confusing to the attacker than the tradi-
tional one; achieving higher security. On the other
hand, any binary message can be encrypted in a DNA
sequence of half its length as will be clarified in the
illustrative example.
3.5 Illustrative Example
Assume the message: Hello and the Key : 2411
The Sender side:
1. Encryption process
(a) Generate 16 random letters using the given key,
(Table 3) will be generated.
(b) Use the key to shuffle the initial values in (Table
4) and (Table 5), where (Table 6) and (Table 7)
will be generated respectively.
(c) Get the Binary form [B] of the message:
0100100001100101011011000110110001101111
(Binary sequence)
(d) Get the English letters sequence by mapping
the position of each 4 bits of B in (Table 6) to
their corresponding positions in (Table 3): Q Z
D A D M D M D V (Original English text)
(e) Perform the playfair cipher process on the En-
glish text: W D G I Z H Z H G Q (Ciphered
text)
(f) Convert the ciphered text to a DNA sequence
using (Table 3) and (Table 7), the resulted se-
quence: CCTATCATGGGTGGGTTCGC (En-
crypted DNA sequence)
Note: the Binary message consists of 40 bits,
encrypted in a DNA sequence of 20 bases. i.e.
half its length.
2. Hiding process
(a) Use a suitable Reference DNA sequence from
NCBI database (NCBI database).
Table 7: 4x4 Shuffled DNA grid.
GT CG CA TG
TA TC GG AA
AT TT CT AG
GC GA CC AC
(b) Hide the Encrypted DNA sequence ”CCTAT-
CATGGGTGGGTTCGC” in the chosen refer-
ence DNA sequence using the substitution pro-
cess mentioned in (Khalifa and Atito, 2012).
(c) The result is a fake DNA sequence.
The Receiver side:
Given the fake DNA sequence, the reference DNA se-
quence and the key.
1. Extraction process
(a) Extract the hidden encrypted DNA sequence,
using the reverse of the substitution process
mentioned in (Khalifa and Atito, 2012).
(b) The result is encrypted DNA sequence: ”CC-
TATCATGGGTGGGTTCGC”.
2. Decryption process
(a) Generate 16 random letters using the given key,
(Table 3) will be generated.
(b) Use the key to shuffle (Table 4) and (Table 5),
(Table 6) and (Table 7) will be generated.
(c) Convert the encrypted DNA sequence to En-
glish letters by mapping their positions in (Ta-
ble 7) to their corresponding positions in (Table
3): W D G I Z H Z H G Q (Ciphered text)
(d) Perform the inverse of playfair cipher process
Q Z D A D M D M D V (Original English text)
(e) Map the positions of the English letters in (Ta-
ble 3) to a binary form in their corresponding
positions in (Table 6)
0100100001100101011011000110110001101111
(f) Convert the Binary form to the original mes-
sage ”Hello”
4 PERFORMANCE ANALYSIS
In this section we discuss the cracking probability as
well as the experimental results on our algorithm im-
plementation
4.1 Cracking Probability
Despite the fact of simplifying the recent DNA-based
playfair cipher algorithm, the cracking probability of
our proposed algorithm remains high and becomes
AnEnhancedDNA-basedSteganographyTechniquewithaHigherHidingCapacity
155
even more confusing to the attacker. In case of
both the recent technique and the proposed one, the
attacker needs 4 types of information to decrypt a
message, Binary representation, Reference DNA, the
Complementary rule (Khalifa and Atito, 2012) and
the ciphering technique. Probability to get the binary
scheme b is:
P(b) =
1
4!
(4)
Since we have 4 DNA bases, the number of possible
binary schemes is 4!.
Probability to get the Reference DNA r:
P(r) =
1
1.6 ∗ 10
8
(5)
Since there exists 1.6 ∗ 10
8
DNA sequences on the
DNA database(NCBI database).
Probability of the complementary rule c is:
P(c) =
1
16
(6)
Therefore the overall cracking probability k is:
P(k) = P(b) ∗ P(r) ∗ P(c) =
1
24 ∗ 1.6 ∗ 10
8
∗ 16
(7)
In case of the ciphering technique, there are 3 as-
pects that make our proposed DNA-based playfair ci-
pher technique stronger than the traditional one.
1. We are using 4x4 grid instead of the traditional
5x5 playfair cipher grid, which means that the at-
tacker might guess a sequence of English letters
which are not in the grid.
2. We are not ciphering plaintext, but we are cipher-
ing the binary form of the plain text. This means
that we can cipher letters, numbers and even punc-
tuation symbols.
3. The Key used in our modified playfair cipher tech-
nique is not restricted to characters only, it can be
of any type since we get the numeral of the key
and use it as a seed value for generating any 16
English letters to be presented by the grid.
All the above new aspects of our proposed mod-
ified DNA-based playfair cipher technique makes it
more robust and its cryptanalysis becomes harder to
break.
4.2 Experimental Results
Experimental results of our proposed DNA-based
Steganography technique was compared with the re-
sults in(Khalifa and Atito, 2012) to confirm the supe-
riority of our proposed algorithm regarding the max-
imum size of bits that can be embedded in the cover
0
10
20
30
40
50
60
145 155 165 175 185 195 205 215 225
Capacity (kb)
Reference DNA sequence Length (bp)
X1000
Current tech.
Proposed tech.
Figure 3: Capacity of the current technique Vs the proposed
one.
media named as the hiding capacity, as well as the
percentage of the maximum hiding capacity needed
to hide the message named as the actual payload and
the execution time. Our algorithm was tested on the
same used 8 benchmarks adopted from the (NCBI
database). As shown in Figure 3, Figure 4 and Figure
5, the x-axis represents DNA sequences with different
sizes used for hiding in terms of base-pairs(bp).
In Figure 3, the y-axis presents the hiding capac-
ity of the 8 reference DNA sequences. Despite the
fact of our proposed technique simplicity, it improved
the hiding capacity by 25% more than the current
technique, since the current technique hides 3/2 bits
per nucleotide, the proposed technique hides 2 bits
per nucleotide; for example a reference DNA of size
149,884 bp can hide a message of length up to 27.44
Kb by using the current technique while the proposed
one can hide up to 36.56 Kb.
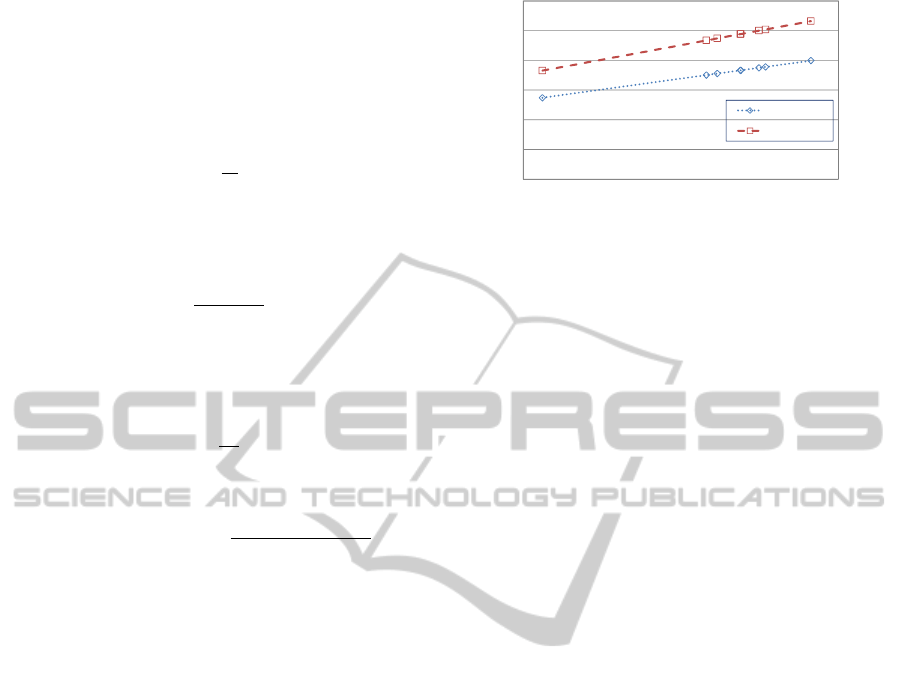
The actual payload of the proposed technique is
compared with that of the current one using a message
of size 10 kb. As shown in Figure 4, the less actual
payload percentage, the more data can be hidden. For
example, the 10 Kb message occupies 26.45% in a
reference DNA of length 206,488 bp using the current
technique, while the same message occupies 19.83%
in the same reference DNA using the proposed tech-
nique. In other words, the proposed technique can
efficiently hide a larger messages in comparison with
the current one, as illustrated by Figure 4. It is worth
to note, that the hiding capacity of the proposed tech-
nique increases by an average rate of 25% in com-
parison with the current technique, while the actual
payload decreases by almost the same rate.
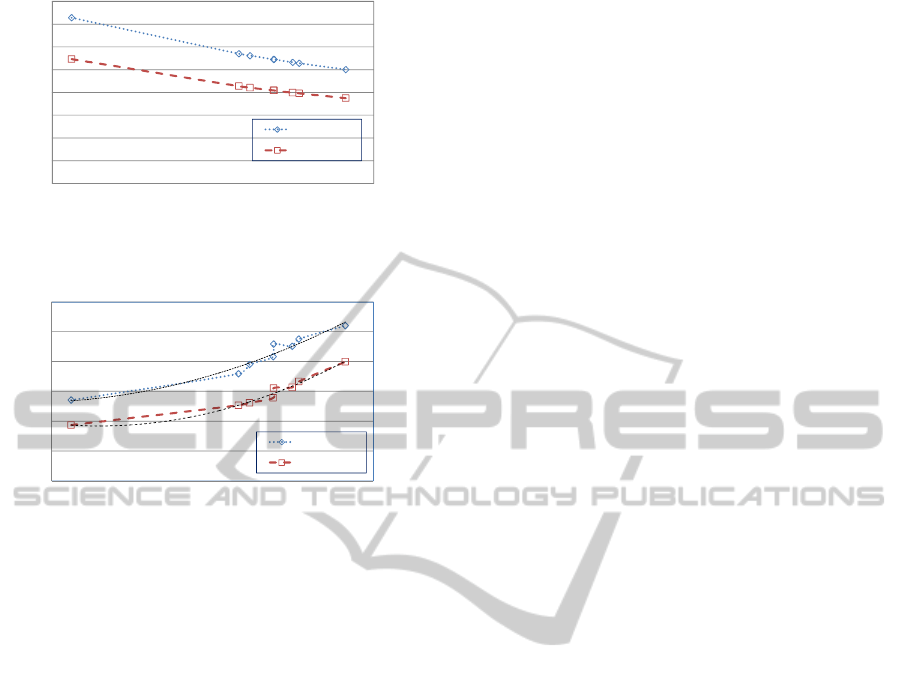
In Figure 5, the y-axis represents the execution
time. The two illustrated curves represent the perfor-
mance of the current hiding algorithm and our pro-
posed one. It is easily noted that the execution time
of the proposed algorithm is significantly less than the
current one as it get rid of the repetitive steps of the
current technique.
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
156
0
5
10
15
20
25
30
35
40
145 155 165 175 185 195 205 215 225
Actual Payload (%)
Reference DNA sequence Length (bp)
X1000
Current tech.
Proposed tech.
Figure 4: Actual payload of the current technique Vs the
proposed one.
0
20
40
60
80
100
120
145 155 165 175 185 195 205 215 225
Time (sec)
Reference DNA sequence Length (bp)
X1000
Current tech.
Proposed tech.
Figure 5: Execution time of the current technique Vs the
proposed one.
5 CONCLUSIONS AND FUTURE
WORKS
This paper proposed a new DNA-based Steganog-
raphy algorithm to achieve a high hiding capacity.
It’s composed of two steps, firstly data encryption
using enhanced DNA-base playfair cipher, Secondly
utilizing the recent substitution technique for hid-
ing. The proposed algorithm enhanced the commonly
used playfair cipher and got rid of the overhead ambi-
guity bits. It proved that a message can be encrypted
in a DNA sequence in half of its length. Moreover,
the hiding capacity of the cover DNA sequence is im-
proved by 25% where each 2 bits are hidden in one
DNA base as a result of the playfair cipher modifi-
cation. Additionally, we enhanced the security and
achieved lower execution time as well. The conducted
experimental studies proved the superiority of our
proposed approach in terms of higher hiding capac-
ity and better time performance in comparison with
the current DNA-based Steganography algorithms.
Using DNA as a medium for Steganography is very
promising, due to the fact of its huge storage capac-
ity. As a future work, we should focus on imitating
the DNA nature by developing algorithms with higher
data hiding capacity.
REFERENCES
Adleman, L. (1994). Molecular computation of solutions to
combinatorial problems. Science 11, 266:1021–1024.
Alberts, B. and Johnson, A. (2008). Molecular Biology of
The Cell. The publishing company, US, 5th edition.
Atito, A., Khalifa, A., and Rida, S.Z. (2012). DNA-Based
Data Encryption and Hiding Using Playfair and In-
sertion Techniques. Journal of Communications and
Computer Engineering, 2:44–49.
Catherine, C., Risca, V., and Bancroft, C. (1999). Hiding
messages in dna microdots. Nature Magazine, 399.
Khalifa, A. and Atito, A. (2012). High-capacity dna-based
steganography. The 8th Int. Conf. on INFOrmatics
and Systems.
Leier, A., Richter, C., Banzhaf, W., and Rauhe, H. (2000).
Cryptography with dna binary strands. BioSystems
57.
NCBI data base. http://www.ncbi.nlm.nih.gov/
Peterson, I. (2001). Hiding in dna. Muse.
Sabry, M., Hashem, M., Nazmy, T., and Khalifa, M. E.
(2010). A dna and amino acids-based implementation
of playfair cipher. Int. Journal of Computer Science
and Information Security, 8.
SAEB, M., EL-ABD, E., and EL-ZANATY, M. E. (2007).
On covert data communication channels employ-
ing dna recombinant and mutagenesis-based stegano-
graphic techniques. pages 200–206. CEA’07 Proceed-
ings of the 2007 annual Conference on Int. Conf. on
Computer Engineering and Applications.
Shimanovsky, B., Feng, J., and Potkonjak, M. (2002). Hid-
ing data in dna. The 5th Int. Workshop on Information
Hiding, 2578:373–386.
Shiu, H., Ng, K., Fang, J., Lee, R., and Huang, C. (2010).
Data hiding methods based upon dna sequences. EL-
SEVIER, 180:2196–2208.
Smith, W. M. and Group, T. T. (2003). Dna based steganog-
raphy for security marking. XIX International Secu-
rity Printers Conference.
Taur, J.-S., Lin, H.-Y., Lee, H.-L., and Tao, C.-W. (2012).
Data hiding in dna sequences based on table look up
substitution. Int. Journal of Innovative Computing, In-
formation and Control, 8:6585–6598.
AnEnhancedDNA-basedSteganographyTechniquewithaHigherHidingCapacity
157
