
Exposome, Health and Biomedical Informatics
An Emerging Discipline and Its Interaction with Current Biomedical Informatics
Guillermo Lopez-Campos
1
, Riccardo Bellazzi
2
and Fernando Martin-Sanchez
1
1
Health and Biomedical Informatics Centre, The University of Melbourne, Berkeley Street, Parkville, Victoria, Australia
2
Dipartamento di Ingengnieria Industriale e dell’Informazione, University of Pavia, Pavia, Italy
Keywords: Exposome, Exposome Informatics, Biomedical Informatics, Health Informatics, Precision Medicine, Big
Data, Small Data.
Abstract: In the last decade we have witnessed the raising of the exposome (the set of a life-long individual
exposures) as an increasingly interesting area and discipline due to its relationship with health. These new
approaches rely heavily in the use of different informatics related methods and are generating new data
types that in the future should be handled by biomedical informatics. This position paper refers to some of
the challenges that are related with these new approaches from a biomedical informatics perspective,
describing the interactions with related disciplines such as bioinformatics, public health informatics and
others. We discuss as well the role of the exposome in bringing new data types that might be handled by
biomedical informatics in the context of Big and small data generated in this approaches and its relationship
with the participatory medicine and how they could influence future health information systems. Finally, we
consider that the current situation of the exposome resembles the early years of genomics, when it was clear
that genomic information had a great potential for health and drove a discussion about how to better
integrate and analyse the most relevant pieces of information for health purposes.
1 INTRODUCTION
Biomedical informatics is a very dynamic discipline
that is being influenced by advances in many other
areas and scientific disciplines. In the last couple of
decades, the revolution in molecular biology, the
advances in the understanding and use of genomic
information and other technological advances, such
as those related with imaging techniques, have
driven major changes in the discipline and have even
shaped subdisciplines such as translational
bioinformatics or imaging informatics.
The relevance of environmental factors for
health has been known since the early dawn of
medicine but it has been more recently when
scientists have had the opportunity to explore and
manage these data in a more comprehensive way. It
has been this capacity of gathering large amounts of
personalised individual exposures what led to the
advent of the concept of the exposome.
The exposome concept was coined almost a
decade ago (CP Wild. 2005) and it was defined as
the life-time set of environmental exposures of an
individual. Therefore the exposome comprises any
environmental exposure of an individual and it
complements the genome in configuring the final
phenotype of an individual and its health status.
The application of the exposome concepts and
studies for health purposes makes it automatically an
area of interest for biomedical informatics. Despite
of the growing interest and relevance of the
exposome in the recent years and the importance of
some of the informatics associated components it
has just recently reached the interest of the
biomedical informatics community. Thus it is
necessary to identify what areas of exposome
informatics are already covered by the current
practice of biomedical informatics and what are the
new challenges that these approaches are posing for
the biomedical informaticians.
In many aspects, from a biomedical informatics
perspective, the exposome could be considered in
the same situation as genomics were a couple of
decades ago, when it started to reach the attention
for its potential health applications and it was
necessary to identify the new challenges that its
integration could pose to develop effective solutions
for medicine.
580
Lopez-Campos G., Bellazzi R. and Martin-Sanchez F..
Exposome, Health and Biomedical Informatics - An Emerging Discipline and Its Interaction with Current Biomedical Informatics.
DOI: 10.5220/0005278405800584
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2015), pages 580-584
ISBN: 978-989-758-068-0
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

2 THE EXPOSOME: AN
INTERSECTION FOR
DIFFERENT DISCIPLINES
The exposome represents a multidisciplinary
challenge involving different scientific disciplines
ranging from the design of new sensors and devices
(engineering) and development of new data analysis
and integration tools (Information Technologies) to
public health studies (Medicine). It is therefore an
area of convergence where different approaches,
methodologies and techniques are combined. An
example of aspects covered could be represented by
the variety of journals where “exposome” related
articles are published (figure 1).
Figure 1: Representation of the orientation of the journals
identified in a Pubmed query (October 2014) using the
term “EXPOSOME”. The majority of them are related
with public health and environment (42%) followed by
clinical journals (30%), Fundamental research (covering
from biology to chemistry) (21%) and finally Technical
Journals (informatics and statistics) (7%).
The exposome serves as well as a good example
for the convergence of the different trends in
medicine that have been defined in the last years
such as molecular, personalised and participatory
medicine but also with other more traditional
approaches such as public health and epidemiology.
Molecular and personalised medicine have a
strong connection with the exposome. Both the
exposome and the personalised medicine approaches
are based in subject’s individual characteristics and
conditions from their very definition and take into
account the individual genomic information. In the
case of molecular medicine, the relationship is based
in the methodologies applied in the studies.
Exposome analyses and methods are strongly
influenced at the molecular level by toxicogenomics,
a discipline related with the study of genomics
effects and responses to toxic substances (Nuwaysir
EF et al. 1999), and make extensive use of “omics”
based techniques and approaches. Probably one of
the best examples are the epigenetic studies that are
supported in the major international activities around
the exposome.
On the opposite side of the spectra from the
molecular and individual approaches we could find
the population health perspective of the exposome
where it makes use of “traditional” environmental
data sources and it is interested on linking exposures
and environmental factors to disease burden. The
aim of this approach is to identify and make sense of
the most relevant exposure factors that are relevant
at the individual level. Similarly to what happened in
genomics and personalised medicine where the
definition of those genomic variants and biomarkers
relevant to a certain disease required from
population studies and genome wide association
studies (GWAS), the exposome requires as well of
these approaches and analogously has developed and
uses the environmental wide association studies
(EWAS) (PM Lind et al. 2013, CJ Patel et al. 2010,
SM Rappaport. 2012).
Finally, the exposome also represents a link with
the increasingly popular movement of participatory
medicine. For a long time exposure data have been
gathered from “general” environmental data
collection tools, such as environmental stations,
public surveys or in a more refined way through
occupational health risks surveys and assessments.
Nowadays the ubiquity of GPS systems in a
multitude of consumer devices has facilitated the
linkage between the individual location and the
exposure in those non-personal environmental data
repositories. Even more, the continuous advances in
technology are developing and creating new
miniaturised devices that can be used to monitor an
increasing range of exposures. Thus the availability
and affordability of these sensors is facilitating its
inclusion in many portable and wearable devices
such as mobile phones (GJ Stahler et al. 2013, MJ
Nieuwenhuijsen et al. 2014) and therefore facilitated
its use by the general public out of the laboratories.
It is therefore, this portability of the new sensors
what has enabled the possibility of capturing some
real time, exposome data that could be then
specifically linked to people’s own health data and
could be use for health management purposes.
3 EXPOSOME RELATED
INFORMATICS
As in many other disciplines informatics has become
a key element for its development, taking advantage
Exposome,HealthandBiomedicalInformatics-AnEmergingDisciplineandItsInteractionwithCurrentBiomedical
Informatics
581

of some of the disciplines it is related with. As it was
discussed previously, the close relationship between
the exposome and toxicogenomics brought the use
of different “omics” technologies and with them as
well a high relevance of bioinformatics approaches
and solutions for data analysis and management.
Additionally, exposome approaches are taking
advantage of the latest advances in systems biology
methodologies and are incorporating and making
extensive use of them. This intensive use of
bioinformatics serves as a link between the
exposome and biomedical informatics. This is so
because bioinformatics approaches related with
health are considered under the umbrella of
biomedical informatics nowadays and due to the
increasing interest in the role of the exposome in
health, the role of bioinformatics applications in this
area should be considered as well. An example of
the high relevance of informatics for the exposome
approaches can be found in the existence of one or
several work packages exclusively devoted to
informatics related aspects in the four major
international exposome projects. (Table 1).
Table 1: Major international projects involving the study
of the Exposome and containing significant informatics
work packages.
Project Name
HELIX - The Human Early Life Exposome. It is a
European funded project focused in the development of
new tools to integrate exposome and childrens’ health
data
http://www.projecthelix.eu/
EXPOSOMICS. It is a European funded projects aiming
to predict individual risk related to the environment.
http://www.exposomicsproject.eu/
HEALS – Health and Environment-wide Associations
based on Large population Surveys. It is a European
funded project.
http://www.heals-eu.eu/
HERCULES – Health and Exposome Research Center:
Understanding Lifetime Exposures. It is an US funded
project.
http://emoryhercules.com/
3.1 The Exposome, Big Data and Small
Data
As it has been previously described one of the most
relevant characteristics of the exposome is that it is
an area where several scientific disciplines converge
and intersect with each other. From a data and
information point of view, this convergence of
multiple disciplines could be translated into a large
diversity of data types and sources of interest that
must be dealt with in the analysis of the exposome.
The different data types that are involved in the
analysis of the exposome range form molecular data
associated either with genomic or pollutant data to
geographical locations. Additionally large data sets
are used in the analysis, coming from a broad variety
of sources (again using as an example the integration
of population scale “omics” studies and large
environmental data sets) that need to be combined.
This combination of large volumes and a large
variety of data types confers exposome data the
category of big data which could be generally
characterised by the four “V’s” (Volume, Variety,
Velocity and Veracity) (A. McAfee et al. 2012)
Even though as it has been discussed before the
exposome is a life-long set of exposures (data)
implying that this is a temporal data collection, The
third V from the big data definition (Velocity) could
be arguably considered as well as part of the
exposome data characteristics due to the continuity
of the measurements that are made during with
many of the devices.
On the other hand the individual and
participatory component of the exposome related
with another increasingly popular topic from an
informatics perspective that is “Small Data”. In
contrast with Big Data, small data come from the
individual digital traces that are created or left
continuously in the use of technology (D. Estrin.
2014). These “Small data” are generated as a
consequence of the use of the portable devices and
self-monitoring practices that could be used to
quantify the individual exposures.
Therefore the exposome represents a multilevel
challenge for the current biomedical informatics
discipline. On one hand the inclusion and
development of exposome data as another element
of biomedical informatics means an extension of an
already existing problem in terms of the needs and
requirements to work in “Big Data” environments
for data analysis and management. In this regard, the
current solutions and approaches would need to be
extended to incorporate these new data sources, and
many of the solutions already in place would just
need to be expanded to accommodate exposome
data.
On the other hand dealing with “Small Data”
represents a new challenge that its starting to be
tackled by biomedical informatics experts. It means
a mutual interest area and therefore it represents a
common area of development where exposome data
should be considered as another source of health
related data and therefore should be incorporated
into future health information systems. The
HEALTHINF2015-InternationalConferenceonHealthInformatics
582
challenge in this case is not only developing new
methods and ways to capture, manage and analyse
these new data sources but also the need to think
about the new data sources and new data types that
are being captured in these projects. An important
and significant difference in this case is that when
dealing with exposome “Small Data” we are talking
in many cases about data that is being generated by a
broad range of devices, that may use or no other
standards than those usually used in biomedical
informatics. Another challenging characteristic of
these new exposome “Small Data” is that they are
captured in a continuous way rather than the
traditional more or less static (snapshots) data
capture processes that have been considered in the
development of health information systems.
4 EXPOSOME, INFORMATION
LEVELS AND PRECISION
MEDICINE
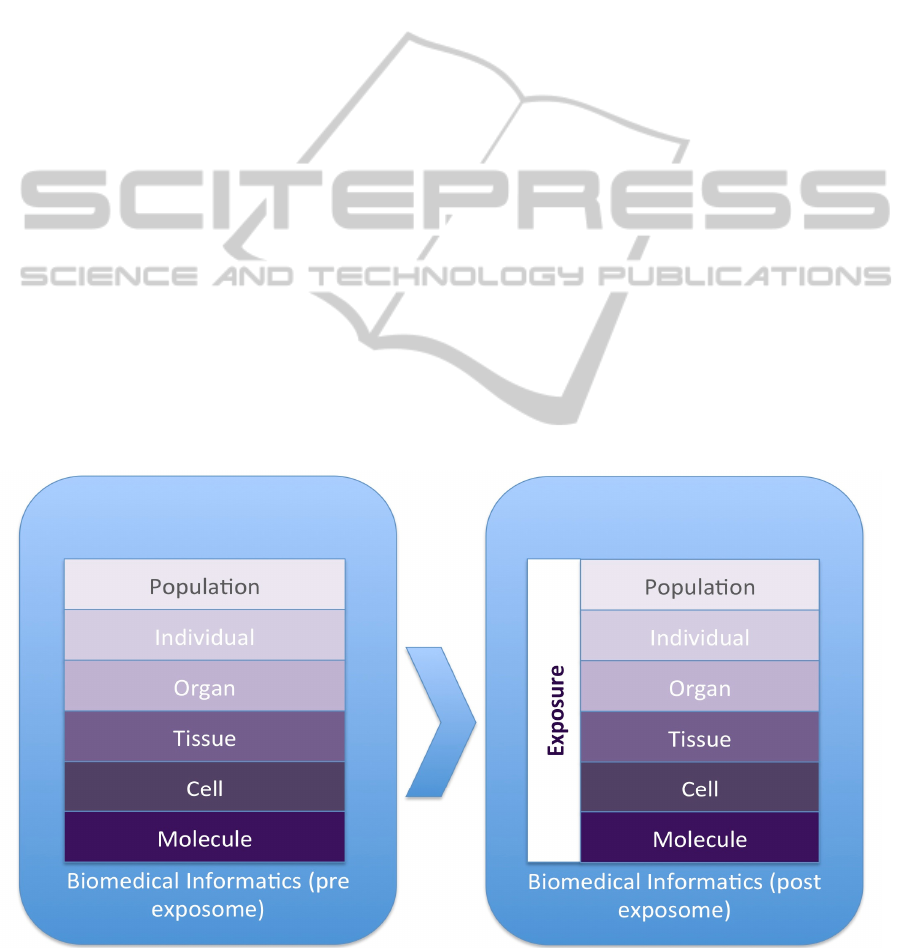
A way to represent the current view of biomedical
informatics is based in the analysis of the different
information levels covered, going from molecule to
population (F Martin-Sanchez et al. 2002). In that
model some of the environmental factors and
elements that are part of the exposome were
scattered across the individual and population levels
but they were not explicitly formulated nor
addressed. With the advent and development of the
exposome, it is possible to develop an additional
vertical layer vertical layer to that model
representing the exposome, interacting with all the
other layers.
The inclusion of this additional layer to the
information and complexity model will create a
more comprehensive model facilitating the path for
the development of biomedical informatics tools and
methods required for precision medicine. Precision
medicine has been defined as the outcome of a
refined and more accurate definition of the disease
based in the integration of different data sources,
coming from improved diagnostics tests (that could
be molecular or genomic) and personal exposure
(exposome), rather than relying on symptoms and
other physical signs (Nat. Acad. Sci. 2011). The
actual implementation of this precision medicine
approach relies heavily in the whole and most
comprehensive body of biomedical informatics as a
discipline that uses biomedical and health data,
information and knowledge.
The actual situation of the exposome informatics
presents some similarities with the situation found a
decade ago when the first genomic approaches were
being developed and there was a need to establish
links between bioinformatics and genomics with
health informatics. Nowadays, the possibility of
Figure 2: Representation of the expanded health information levels that are covered by biomedical informatics where the
new exposure data should be considered as another layer additional to those previously covered by health and biomedical
informatics.
Exposome,HealthandBiomedicalInformatics-AnEmergingDisciplineandItsInteractionwithCurrentBiomedical
Informatics
583

effectively capturing and exploiting exposure
information is promising for a better classification
and understanding of the pathological processes and
therefore it requires a response from the biomedical
informatics to effectively incorporate this data,
information and knowledge with the aim of
improving human health and clinical practice.
REFERENCES
Estrin, D., 2014. Small data, where n=me.
Communications of the ACM, 57 :4, 32-34.
Lind, P. M., Riserus, U., Salihovic, S., Bavel, B. & Lind,
L. 2013. An environmental wide association study
(EWAS) approach to the metabolic syndrome. Environ
Int, 55, 1-8.
Martin-Sanchez, F., Maojo, V. & Lopez-Campos, G.
2002. Integrating genomics into health information
systems. Methods Inf Med, 41, 25-30.
McAfee A, Brynjolfsson E., 2012. Big data: the
management revolution. Harvard Business Review,
90:60-66, 68, 128.
National Academies of Sciences. Toward Precision
Medicine: Building a Knowledge Network for
Biomedical Research and a New Taxonomy of
Disease. Washington (DC).
Nieuwenhuijsen, M. J., Donaire-Gonzalez, D., Foraster,
M., Martinez, D. & Cisneros, A. 2014. Using personal
sensors to assess the exposome and acute health
effects. Int J Environ Res Public Health, 11, 7805-19.
Nuwaysir, E. F., Bittner, M., Trent, J., Barrett, J. C. &
Afshari, C. A. 1999. Microarrays and toxicology: the
advent of toxicogenomics. Mol Carcinog, 24, 153-9.
Patel, C. J., Bhattacharya, J. & Butte, A. J. 2010. An
Environment-Wide Association Study (EWAS) on
type 2 diabetes mellitus. PLoS One, 5, e10746.
Rappaport, S. M. 2012. Biomarkers intersect with the
exposome. Biomarkers, 17, 483-9.
Stahler, G. J., Mennis, J. & Baron, D. A. 2013. Geospatial
technology and the "exposome": new perspectives on
addiction. Am J Public Health, 103, 1354-6.
Wild, C. P. 2005. Complementing the genome with an
"exposome": the outstanding challenge of
environmental exposure measurement in molecular
epidemiology. Cancer Epidemiol Biomarkers Prev, 14,
1847-50.
HEALTHINF2015-InternationalConferenceonHealthInformatics
584
