
Archetypes Development in Electrophysiology Domain
Electroencephalography as a Personal EHR System Module
V´aclav Papeˇz
1,2
and Roman Mouˇcek
1,2
1
Department of Computer Science and Engineering, University of West Bohemia, Pilsen, Czech Republic
2
NTIS New Technologies for Information Society, University of West Bohemia, Pilsen, Czech Republic
Keywords:
Archetype, Template, EHR, OpenEHR, Neuroinformatics, Ontology, Electrophysiology,
Electroencephalography.
Abstract:
The work presents the concept of a new personal electronic health record (EHR) system. The system is
based on openEHR standards/framework. A fundamental part of openEHR is domain description by two layer
modelling - reference data models and archetypes. Archetypes are building blocks of the EHR system, which
provide structure and semantics for stored data. As common ontologies and terminologies, even archetypes
are based on reusability. Although the public archetype repositories (clinical knowledge managers, CKM)
contain hundreds of archetypes from various domains, the electrophysiology domain is not described yet. The
work is focused on the development of the archetypes dealing with the electroencephalography domain.
1 INTRODUCTION
Neuroinformatics research group at the University
of West Bohemia is focused on the development
and integration of software tools and hardware
devices for EEG/ERP (Electroencephalography /
Event-Related Potentials) research, design and
applications of signal processing methods, and design
and implementation of EEG/ERP experiments. The
EEG/ERP Portal (EEGBase) (Jezek and Moucek,
2012) , as one of the group projects, is a software
solution for storing, sharing and analysing data
from neurophysiological experiments. Recently
we have started to expand into other fields
of bioinformatics (ECG, electrocardiography) and
assistive technologies (smartphone applications using
smartphones sensors). The ability to measure and
gather data describing personal health conditions
and activities represents a challenging task; data are
heterogeneous and they require a set of metadata
to clarify their meaning. There are a lot of
applications focused on sportsmen’ training, users’
diet or patients’ sleep quality. However, it is difficult
to analyse data across these applications and look for
non-trivial consequences. Opposed to one-purposed
solutions, there are complex electronic health record
systems (EHR), used in national health care systems,
hospitals or at least doctor’s offices. These systems
are not flexible, because of strict respect of national
standards and orientation on physicians. Therefore,
an idea of personal flexible EHR has arisen.
Currently, mobile devices and sensors are able
to measure basic biological functions. Beside
respiration sensors, heart-rate sensors, pocket-size
ECG, even simplified electroencephalogram is
now easily available via simple, one-electrode,
cordless headsets. Moreover, applications for
brain training, compatible with mobile EEG
headsets, exist as well. Brain data gathering (in a
simplified form) is now easy to do as cardio data
gathering. EEG (unlike e.g. ECG) has a limited
representation in terminologies, ontologies or EHR’s
standards/frameworks. OpenEHR (Beale et al., 2006)
as one of such frameworks, was chosen for the
personal EHR system for its user-centred approach.
It provides the concept of two-layer modelling based
on archetypes and reference models. Since the EEG
domain is not represented in the public archetype
knowledge base, new archetypes describing this
domain should be proposed.
Know-how from the electrophysiology domain,
absence of EEG archetypes and rising accessibility of
EEG devices in households are the main reasons why
the EEG domain is chosen as the first use case in a
new system.
The paper is organized as follows. Section 2
presents the state of the art. Section 3 specifies the
concept of universal personal EHR system. EEG
611
Papež V. and Mou
ˇ
cek R..
Archetypes Development in Electrophysiology Domain - Electroencephalography as a Personal EHR System Module.
DOI: 10.5220/0005282806110616
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2015), pages 611-616
ISBN: 978-989-758-068-0
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)
metadata are described in Section 4. The 5
th
section describes proposed archetypes and templates.
Concluding remarks are given in Section 6.
2 STATE OF THE ART
Generally, EHR systems are designed in three ways
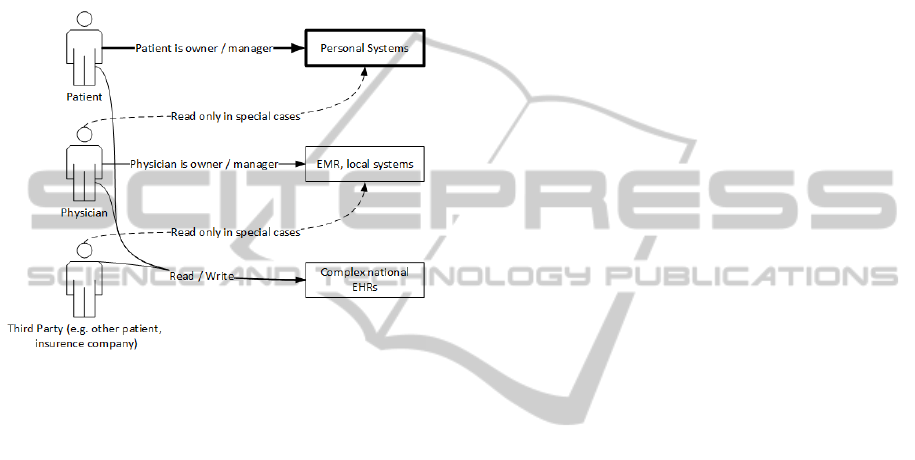
(Figure 1).
Figure 1: Users of EHR systems.
• Personal EHRs: users’ personal access only;
mostly independent of national standards;
• EHR systems oriented on physicians: physicians’
access only; dependent on national standards;
• The combination of previous two concepts:
physicians’ and patients’ read and/or write access;
dependent on national standards.
Besides the general concept, many standards,
methodologies and architectural patterns exist.
Standards are often complementary. Some of the
most respected international standards are:
• HL7 (Dolin et al., 2006)A set of international data
transfer standards, guidelines and methodologies.
Fundamental parts are conceptual standards
(e.g. RIM), document standards (e.g. CDA),
messaging standards and application standards
(e.g. CCOW).
• CEN/ISO 13606 (Sato and Luhn, 2007) A
European standard defining an information
architecture of EHR. It is based on the generic
information models. The architecture is proposed
to be mapped to HL7 v3.
• openEHR: An open standard which brings
two layer modelling methodology (the general
concept is formed by an information/reference
model and the specified item is formulated by
archetypes). It is based on ISO 13606. Its models
describe demographics, services, clinical content
and clinical workflows. The clinical content is
modelled by archetypes and templates.
Apart from an architectural standard, unified
terminology for data description is necessary
as well. One of the biggest worldwide used
multilingual terminology for many medical domains
is SNOMED CT (Stearns et al., 2001). However,
more suitable ontologies/terminologies exist for
the EEG domain. The following list represents
significant ontologies/terminologies in (neuro)
electrophysiology.
• Open Metadata Markup Language (OdML)
(Grewe et al., 2011)) (odML consist of
domain independent data transport format
and electrophysiological terminology)
• Neural Electro Magnetic Ontologies NEMO
(Frishkoff et al., 2009)
• Ontology for describing Experimental
Neurophysiology OEN (Bruha et al., 2013)
Before specific terminology/standard/framework
selection is presented, the general concept and target
users of the EHR system will be described.
3 CONCEPT OF UNIVERSAL
EHR SYSTEM
3.1 General Concept
The general concept of our EHR system is based
on a continuous gathering of personal bio data.
Gathered data are stored, browsed, analysed and
eventually shared. All objectives are focused on one
user/patient/sportsman, not on a physician. The main
goal is to develop a knowledge base and software tool
which will be extendable with new features / clinical
contents / analytic methods as implemented plugins.
The user will have
• immediate access to short time health statistics
• ability to compute long term health statistics
• ability to view trends of his/her clinical contents
and their dependencies
The features mentioned above can reveal potential
health problems sooner than they become serious (or
at least increase the motivation for a healthy lifestyle).
The target group of users, in fact, can be anyone,
but the following three main groups / use-cases are
supposed (not disjunctive):
HEALTHINF2015-InternationalConferenceonHealthInformatics
612
• healthy adults: everyday or occasional monitoring
of their health condition, better life style
motivation, long-term disease prevention
• seniors: everyday monitoring, assistive
technology/function, easy data transport to
their physicians
• active people, sportsmen: motivation,
performance monitoring, training / diet plans
3.2 Technical Background
According to Figure 1, our system belongs to the
personal EHR systems. This is one of the main
reasons, why openEHR, which is more user-centred
than HL7, is used. Other concepts were not taken into
considerationbecause of their insufficientapplication.
OpenEHR is based on a two layer modelling. The
first layer is a reference/information model (RM).
It is an abstract description of a piece of medical
domain (e.g. observation, instruction or entry) and
it contains general data structure. An archetype is
a small unit built above the reference model which
describes a particular part of domain (e.g. blood
pressure observation or instructions for transfusion).
It adds attributes, so-called datapoints, to the RM
structure and adds more restrictions (e.g. ranges,
cardinality, etc.) and terminology.
Archetypes describe a domain (or its parts) in the
most general way. Particular applications then could
need only the subsets of the attributes with more
restrictions. Therefore, there are templates.
Terminologies play a big role in archetype
modelling. Good practise is to use some proved
terminology/ontology for new archetypes (e.g.
SNOMED CT). For electrophysiology archetype
modelling, odML terminology is used. There are
three reasons for this decision:
• odML terminology specializes on
electrophysiology and provides terms more
suitable for EEG experiments than SNOMED CT
does.
• OEN as the complex ontology for EEG
experiments is still under development.
• EEGBase is going to integrate odML terminology.
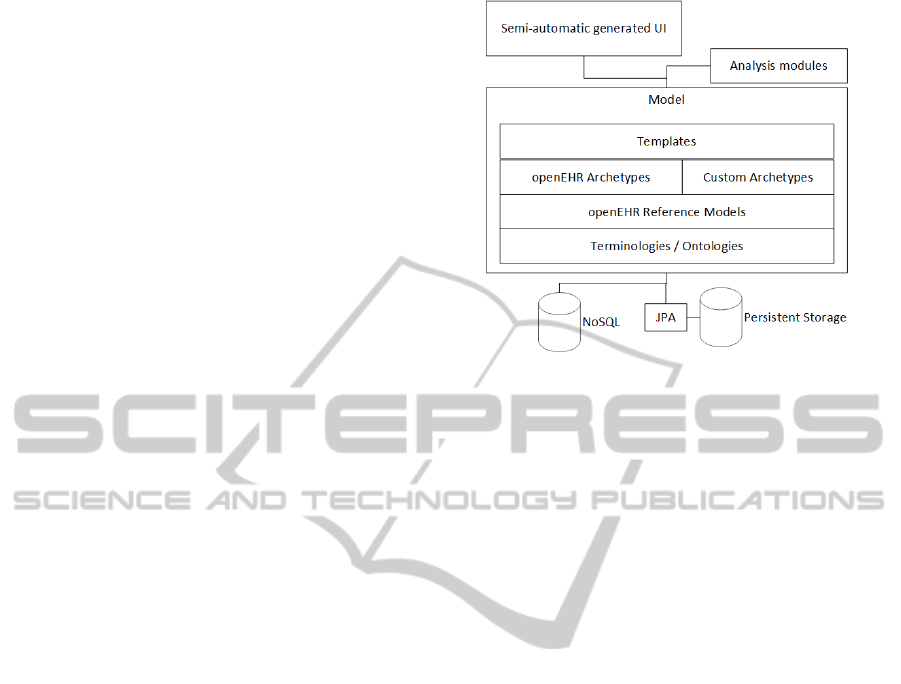
3.3 Architecture
The system architecture is designed to be most
resistant to changes in archetypes/templates.
Archetypes can be seen as a centre of the system
and the parts of other application layers adapt to
it (persistent storage, user interface). A persistent
Figure 2: Basic architecture.
storage bellow the archetypes/templates is realized
via JPA (Java Persistent API) and no-SQL database.
A web based interface uses semi - automatic
UI generation (Figure 2). The goal is to create
easy-install modules by simple adding the archetypes
(in ADL; archetype definition language (Beale et al.,
2005)), templates (in XML or OET format) and
alternatively additional information in configuration
file (e.g. mapping details for JPA).
3.4 Development
Development of the system is divided into four
different phases.
• Archetype and templates modelling
• Mapping between models (archetypes, templates)
and persistent storage
• Semi-automatic generated UI
• Analytic functions and plugins
Recently, the first branch is focused on the EEG
domain (more in sections 4 and 5). The branch
dealing with the analytic modules is postponed until
a reasonable amount of data will be gathered. All
branches and development time estimates are shown
in Figure 3.
Hereafter, the text is focused on the first
branch, i.e. the EEG domain archetypes/templates
development.
4 EEGBase AND odML
METADATA SET
Archetype development has started from an original
set of metadata taken from the EEGBase. It
ArchetypesDevelopmentinElectrophysiologyDomain-ElectroencephalographyasaPersonalEHRSystemModule
613
Table 1: EEGBase Attributes to ODML Terminology.
EEGBase odML Terminology EEGBase odML Terminology
Person
Type: String Type: String
Given name: String
FirstName: String Description: String Description: String
Surname: String FullName: String
Electrode
Date of birth: Date Birthday: Date Location: List ElectrodeLocation: List
Gender: Char
Gender: Char Impedance: Number Impedance: Float
Email: String E-mail: String System: List Usage: List
Phone Number: String PhoneNumber: String
Type: List Type: List
Laterality: Char Laterality: Char
Scenario/Protocol
Education Level: List EducationLevel: List Title: String Name: String
Experiment
Length: Number Duration: float
Start time: Date Recording/Start: Datetime Description: String Description: String
End time: Date Recording/End: Datetime Name: String Author: String
Temperature: Number
Environment/RoomTemperature: String
Software
Environment note: String Environment/Description: String Title: String
Name: String
Quality: String Dataset/Quality: String Description: String
Description: String
Movie: String Stimulus/Movie: String
Weather/Environment
Note: String Description: String Weather: String Weather: String
Stimulus
RoomTemperature: String RoomTemperature: String
Description: String Description: String AirHumidity: String AirHumidity: String
Type: List Type: List
Description: String Description: String
Hardware Disease
Title: String Model: String Description: String Subject.HealthStatus: String
Figure 3: Development chart.
was consulted with domain experts from other
research groups and from hospitals. Since EEGBase
terminology has no standard yet, it was merged with
odML terminology within the project NIX (NIX,
2014). The resulting set of metadata was used for
the archetypes development. Table 1 shows the list of
EEGBase metadata set and its odML alternatives and
it represents the initial attribute list for the templates
design. For the further version of archetypes, OEN is
expected to be used.
5 ARCHETYPES AND
TEMPLATES IN
ELECTROPHYSIOLOGY
5.1 Proposals of New Archetypes
One of the key property of reference models and
archetypes is their reusability. Clinical Knowledge
Managers (CKM) of openEHR and NEHTA
(Australian National E-Health Transition Authority)
(May, 2004) provide hundreds of standardized
archetypes. Suitable existing archetypes were
located and reused from openEHR’s - Device
(Heather, 2010)), Device details (McNicoll, 2010)
and Environmental Conditions (Heather, 2008).
Device and Device details archetypes are used for
Hardware metadata.
The environmental conditions archetype
(archetype available in openEHR CKM; Figure
4) represents a part of the weather attributes.
For the new attributes (Weather and Environment
description) a new archetype Environment with the
HEALTHINF2015-InternationalConferenceonHealthInformatics
614
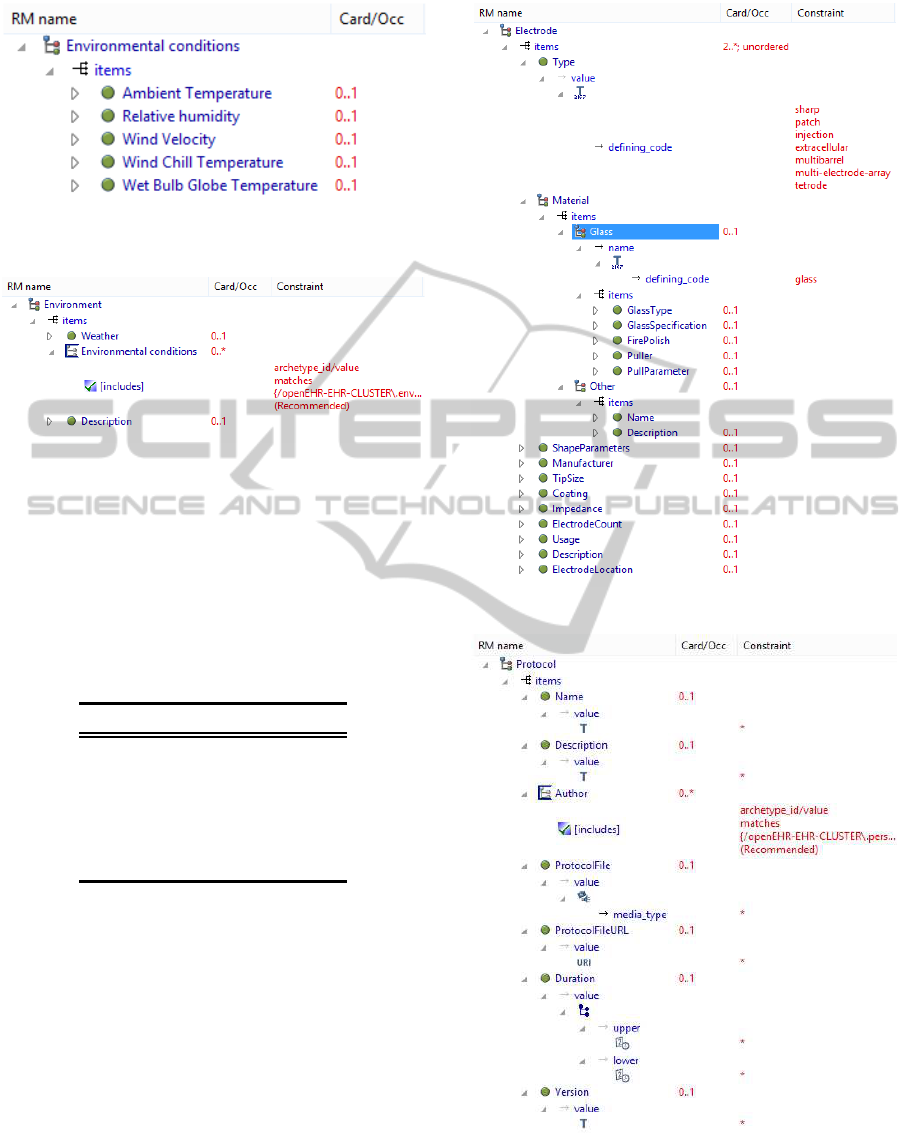
Figure 4: Environmental conditions archetype (Heather,
2008).
Figure 5: Environment archetype; Weather and Description
datapoints contain textual values.
slot for Environmental conditions archetype was
designed (Figure 5).
Table 2 shows all new proposed archetypes and
their reference models. Except for the Experiment
archetype, all archetypes are designed as clusters of
RMs Entries.
Table 2: New archetypes and their reference models.
Archetype Reference Model
Experiment Observation
Stimulus
Cluster of Entries
Software
Cluster of Entries
Scenario
Cluster of Entries
Electrode
Cluster of Entries
Environment
Cluster of Entries
5.2 Development
All proposed archetypes are currently in a draft
state and they are going to undergo team review.
As it was mentioned before, the first version of
archetypes uses odML terminology. Templates than
reduce the number of archetype datapoints to the
original EEGBase metadata set. A few examples
of a new archetypes follow. Electrode description
in odML terminology provides special attributes for
glass electrode. This attribute branching is solved
by the construct dependencies. Electrode archetype
(Figure 6) solves the branching in case of glass
material by new nested cluster.
The protocol archetype (Figure 7) shows how
Figure 6: Electrode archetype.
Figure 7: Protocol archetype.
the protocol is defined in odML terminology. This
protocol definition is based on EEGBase metadata.
Rest of the templates and archetypes can be found at
ArchetypesDevelopmentinElectrophysiologyDomain-ElectroencephalographyasaPersonalEHRSystemModule
615

(https://github.com/NEUROINFORMATICS-GROUP
-FAV-KIV-ZCU/sehr.
The second version of archetypes is planned to use
more complex neurophysiological ontology. The next
archetype lifecycle states (candidate and published)
will come with the second version.
6 CONCLUSION
The work describes a concept of a new EHR system
based on openEHR. It is focused on measured
subjects/users, not on physicians. Its main idea is
to provide an easily extensible system, which is able
to store daily measured medical data. These data
can be further analysed and shared. The openEHR
concept leads to the new archetype proposals. The
electroencephalographydomain was the first use-case
and new archetypes derived from the EEGBase data
structure were proposed. Moreover, archetypes
respect odML terminology.
Archetypes are currently in the lifecycle state
Author’s draft. The future work will focus on
testing these archetypes. With tested archetypes
an EEG plugin for personal EHR system will be
developed. After that, new versions of archetypes
will be designed and implemented; those archetypes
will be based on more complex electrophysiological
ontology OEN. The future development of the EHR
system itself is shown in Figure 3.
ACKNOWLEDGEMENT
The work was supported by the European Regional
Development Fund (ERDF), Project ”NTIS - New
Technologies for Information Society”, European
Centre of Excellence, CZ.1.05/1.1.00/02.0090
and UWB grant SGS-2013-039 Methods and
Applications of Bio and Medical Informatics.
REFERENCES
Beale, T., Heard, S., Kalra, D., and Lloyd, D. (2005).
Archetype definition language (adl). OpenEHR
specification, the openEHR foundation.
Beale, T., Heard, S., Kalra, D., and Lloyd, D. (2006).
Openehr architecture overview. The OpenEHR
Foundation.
Bruha, P., Papez, V., Brandowski, A., Grewe, J., Moucek,
R., Tripathy, S., Wachtler, T., and Le Franc,
Y. (2013). A formal ontology for describing
experimental neurophysiology. Neuroinformatics.
Dolin, R. H., Alschuler, L., Boyer, S., Beebe, C., Behlen,
F. M., Biron, P. V., and Shvo, A. S. (2006). Hl7
clinical document architecture, release 2. Journal
of the American Medical Informatics Association,
13(1):30–39.
Frishkoff, G., LePendu, P., Frank, R., Liu, H., and Dou, D.
(2009). Development of neural electromagnetic
ontologies (nemo): ontology-based tools for
representation and integration of event-related
brain potentials. In ICBO09: Proceedings of the
international conference on biomedical ontology,
pages 31–34. Citeseer.
Grewe, J., Wachtler, T., and Benda, J. (2011). A bottom-up
approach to data annotation in neurophysiology.
Frontiers in neuroinformatics, 5.
Heather, L. (2008). openehr/adl-archetypes.
Heather, L. (2010). openehr/adl-archetypes.
Jezek, P. and Moucek, R. (2012). System for eeg/erp
data and metadata storage and management. Neural
Network World, 22(3):277–290.
May, L. (2004). The national e-health transition authority
(nehta). The HIM journal, 34(1):19–20.
McNicoll, I. (2010). openehr/adl-archetypes.
NIX, G.-N. (2014). G-node/nix.
Sato, L. and Luhn, K. (2007). Cen/iso 13606 pilot study
final report. NHS Connecting for Health [Online].
Stearns, M. Q., Price, C., Spackman, K. A., and Wang,
A. Y. (2001). Snomed clinical terms: overview
of the development process and project status. In
Proceedings of the AMIA Symposium, page 662.
American Medical Informatics Association.
HEALTHINF2015-InternationalConferenceonHealthInformatics
616
