
CereVA
Visual Analysis of Functional Brain Connectivity
Michael de Ridder
1
, Karsten Klein
2
and Jinman Kim
1
1
School of IT, The University of Sydney, Sydney, Australia
2
School of IT, Monash University, Melbourne, Australia
Keywords:
Visual Analysis, Resting State Networks, Graph Layout, Interactive Visualization.
Abstract:
We present CereVA, a web-based interface for the visual analysis of brain activity data. CereVA combines 2D
and 3D visualizations and allows the user to interactively explore and compare brain activity data sets. The
web-based interface combines several linked graphical representations of the network data, allowing for tight
integration of different visualizations. The data is presented in the anatomical context within a 3D volume
rendering, by node-link visualizations of connectivity networks, and by a matrix view of the data. In addition,
our approach provides graph-theoretical analysis of the connectivity networks. Our solution supports several
analysis tasks, including the comparison of connectivity networks, the analysis of correlation patterns, and the
aggregation of networks, e.g. over a population.
1 INTRODUCTION
Unraveling the mysteries of the brain’s inner work-
ings is one of the great challenges in biomedical re-
search today. Recent efforts including the Human
Connectome Project, the Human Brain Project (Van
Essen et al., 2013; HBP, 2014) and the BRAIN ini-
tiative underline the importance of this task. A core
component of these initiatives is to study the patterns
that are evident in the brain activity correlation data,
known as functional connectivity networks. Differ-
ences in the networks between individuals over time,
and between individual networks and aggregated net-
works among the healthy population or groups of pa-
tients can be examined to detect and understand the
alterations related to disorders, e.g. in the context of
Alzheimer’s disease (Greicius et al., 2004). The anal-
ysis of such functional connectivity networks can fos-
ter the detection and understanding of brain activity
patterns (Smith et al., 2013).
Brain activity is commonly measured using neu-
roimaging scanners of functional magnetic resonance
imaging (fMRI). fMRI data is used to first derive
time-course data, from which correlation matrices are
calculated, e.g. (van den Heuvel and Pol, 2010; Smith
et al., 2013). These matrices contain activity correla-
tion values between regions of the brain that are either
defined by a brain parcellation, based on the anatomy,
or by the resolution of the imaging scan, e.g. the vox-
els of an fMRI session (Destrieux et al., 2010). Func-
tional networks are not directly given by this raw data,
but need to be modelled from the correlation matri-
ces. However, a large part of the correlation data is
regarded as noise as they are either redundant or does
not contain meaningful correlation. It is not obvi-
ous how to select the relevant entries from the matri-
ces where each individuals have unique correlations;
a constant ‘threshold value’ might not be sufficient
to cover all biologically significant correlations (Lee
et al., 2012). The need for interactive selection of the
threshold raises opportunities for the application of
visual analysis concepts, where modelling a weighted
graph that captures the main connectivity patterns in
the functional networks then permits graph theoreti-
cal and statistics methods to be employed for analysis
(Onoda and Yamaguchi, 2013; Wang et al., 2010).
While resting state neural networks, which oc-
cur when no conscious activity is being performed,
exhibit several specific properties that can be calcu-
lated based on the network topology, e.g. (Bullmore
and Sporns, 2009; Wang et al., 2010; Chang et al.,
2013), visualization of such networks and the under-
lying data is an important step in facilitating analysis.
Visualization allows the researcher to create insight
by exploiting the humans expertise and ability to spot
patterns, and can also help to cope with the impreci-
sion of the network model and the influence of noise
and uncertainty in the data. Research into the anal-
ysis of neural activity data often includes visualiza-
tion of anatomical and functional information, such as
brain and brain region depiction, e.g. (Nowke et al.,
2013), or neural network visualization, e.g. (Sorger
131
de Ridder M., Klein K. and Kim J..
CereVA - Visual Analysis of Functional Brain Connectivity.
DOI: 10.5220/0005305901310138
In Proceedings of the 6th International Conference on Information Visualization Theory and Applications (IVAPP-2015), pages 131-138
ISBN: 978-989-758-088-8
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

et al., 2013). An investigation into these visualiza-
tions by Margulies et al. (Margulies et al., 2013) ar-
gues that the best views intuitively combine network
and anatomical data without cluttering the anatomi-
cal context, which could potentially present mislead-
ing information. This holds, in particular, when sup-
porting a guided interactive exploration of activity
data. The visualization can be accompanied by a net-
work analysis that helps to identify and exploit the
structural characteristics of the network (Wang et al.,
2010). The overarching goals of these works are
comparison across the population and between indi-
viduals to derive global patterns and variations, e.g.
caused by disease, and to analyze the spatiotemporal
dynamics in order to better understand the structure-
function relationship underlying the connectivity pat-
terns. Faithful network representations are thus re-
quired that convey patterns, as well as strong, clear
links to the anatomical context.
We present a Cerebral Visual Analytics tool,
CereVA, a preliminary approach that facilitates the
analysis of brain activity data by providing a web-
based visual interface for interactive exploratory anal-
ysis of the data. The interface combines several
linked graphical representations of the network data,
allowing for tight integration of different visualiza-
tion views. The data is presented in the anatomi-
cal context within a 3D volume rendering, by node-
link visualizations of connectivity networks, and by
a matrix view of the data. In addition, our approach
provides graph-theoretical analysis of the connectiv-
ity networks. Our solution supports several analysis
tasks, including the comparison of connectivity net-
works, the analysis of correlation patterns, and the
aggregation of networks, e.g. over an aggregated pop-
ulation.
2 APPLICATION AND
VISUALIZATION TASKS AND
CHALLENGES
Important analysis tasks for resting state activity data
include:
A) identification of relevant differences between in-
dividual or aggregated networks
B) identification of significant patterns in a set, e.g.
across the whole population
C) identification of network classes, e.g. for healthy
individuals, disorders and behavioural patterns
D) investigating changes over time, e.g. during de-
velopment (BSA, 2014) or due to aging
These tasks lead to challenges including:
i) modelling weighted graphs from the correlation
matrix that reflect the main correlation character-
istics
ii) modelling aggregated or aligned networks that
represent a whole set of networks, e.g. the ag-
gregate across the population, covering the overall
characteristics well without a large deviation
iii) supporting tasks A and B by intuitive interactive
visualizations for comparison to show dissimilar-
ities and similarities
iv) supporting task C by including automated analy-
sis in the visual interface, e.g. classification and
clustering
v) supporting task D by allowing the analysis of time
series data.
Therefore an intuitive visualization that combines
both functional and anatomical contexts is suitable to
facilitate an exploratory analysis of functional con-
nectivity networks. Several methods of combining
network visualization with imaging data are have
been investigated in recent research (B
¨
ottger et al.,
2014; Sorger et al., 2013; Hagmann et al., 2008).
In addition, there is evidence that matrix-based visu-
alizations can be advantageous for several compari-
son tasks (Alper et al., 2013). Matrices are also well
suited for use in small multiples visualizations, which
can be exploited to represent temporal dynamics or
for comparison tasks. Thus incorporating a matrix-
based view into a visualization approach can facili-
tate comparison as long as it is smoothly integrated
into the interaction concept and does not increase the
complexity of the visualization.
3 CereVA
As the underlying mechanisms of brain function and
the impact of diseases are not yet fully understood, an
interactive visual analysis process is proposed to facil-
itate knowledge discovery. This approach combines
the strengths of the human expert to discover pat-
terns and to exploit existing knowledge with the com-
putational power of automated analysis and guided
navigation. CereVA provides the researcher with an
interactive visual interface that supports such a pro-
cess, combining visualizations of the activity data, the
anatomy and results from analysis. We aim to provide
a system of tools that addresses the tasks described
in Section 2, where we focus on tasks A, B, and C.
CereVA’s design was developed according to require-
ment analysis that included consultations with, and
IVAPP2015-InternationalConferenceonInformationVisualizationTheoryandApplications
132

feedback from, potential users, including an expert.
We provide the implementation of our concept in a
web-based application to allow users anywhere, any-
time access.
While questions asked in the investigation of
disease-related anomalies cannot be answered purely
by automated analysis, the results of the analysis can
still be used to guide the interactive exploration of
the data. The visualizations in our system are there-
fore enriched with the functional activity data and in-
formation derived from it. For the exploratory anal-
ysis, we facilitate interactive navigation through the
given data sets, in particular selection and compari-
son of networks of interest. Our interaction concept
is thus designed to support the following workflow: a
user can either visualize a single network or compare
networks with each other, wherein the networks are
either individual subject networks or aggregated net-
works that are derived from a set of networks, e.g. the
network created by computing the average connectiv-
ity value for each pair of nodes. Results from data
and network analysis are shown to the user and also
mapped onto the network and anatomical visualiza-
tions. By filtering and selecting, the user can investi-
gate the structure and patterns of the activity data.
The visualization components, the underlying
data, the view enrichment, and the interaction concept
are described in the following sections.
3.1 Data Processing and View
Enrichment
Providing the results of an automated data analysis
and mapping these results in an intuitive way onto the
visualizations is important for supporting an efficient
interactive exploration of the activity data.
Network Definitions. To cope with challenges i)
and ii), our concept includes both automated network
creation, and networks based on thresholds interac-
tively defined by the user. Three different types of
networks are used in our approach, data, aggregation,
and comparison networks. All networks contain the
same nodes, brain regions specified in the input data.
A data network is simply a network modeled using
a single correlation matrix to create the edges. An
aggregation network provides a population view —
we create networks that represent the median and the
average pairwise correlation values. A comparison
network is modeled based on differences and similar-
ities between two individuals, including aggregation
matrices.
Threshold Guidance. In general a constant thresh-
old value will not be sufficient for the decision of
which correlations are significant, as this might vary
across different experiments, individuals, or even time
points for one individual. Useful generic models that
best support a specific task or question at hand are
however not known, and thus our system guides the
user in the decision for a correlation value interval
of interest. In order to support the user in selecting
a threshold, the distribution of correlation values is
shown as a heat map. The user can select a correla-
tion value interval of interest using a slider element
on which the above information is displayed. Stan-
dard characteristics such as connectivity, number of
edges and density are calculated for a number of user-
selectable threshold values and can be used guide se-
lection of which correlation threshold might be suit-
able to model the network.
Network Comparison. For correlation values
within the interval an edge is then modelled in the
network, see Figure 1 top right. In a basic implemen-
tation for the comparison of two individual networks,
we create a difference network with an edge mod-
elled if the difference of correlations between two
nodes is between user-defined threshold values. By
using this simple model, the dissimilarities between
networks are emphasized. As an alternative, when
using the circular layout, we include information
on both similar and dissimilar correlation values. A
high value in either network with a small correlation
difference to the other network leads to the creation
of an edge in the comparison network, whereby the
visual representation is altered to show the different
characteristics, as shown in Fig. 3. With this model,
both common patterns and outlier correlations in both
networks are shown, however, the resulting network
is also more dense and thus can be more difficult to
interpret.
Graph Analysis. Analysis methods can be classi-
fied into three major categories, the analysis might be
based on the raw data, taking into account all correla-
tion values, might use the weighted graph modeled
from the data, for example after some user-defined
correlation value threshold is taken into account, or
at the most abstract level, only the graph’s topology
might be used. In particular for the graph-based vari-
ants a large number of measures exist for analyzing
the characteristics. These can be global values, for
example the characteristic path length, or local val-
ues, such as the node strength. As we have a labeled
network with annotations for the different nodes and
their brain regions, we assume that purely topological
CereVA-VisualAnalysisofFunctionalBrainConnectivity
133
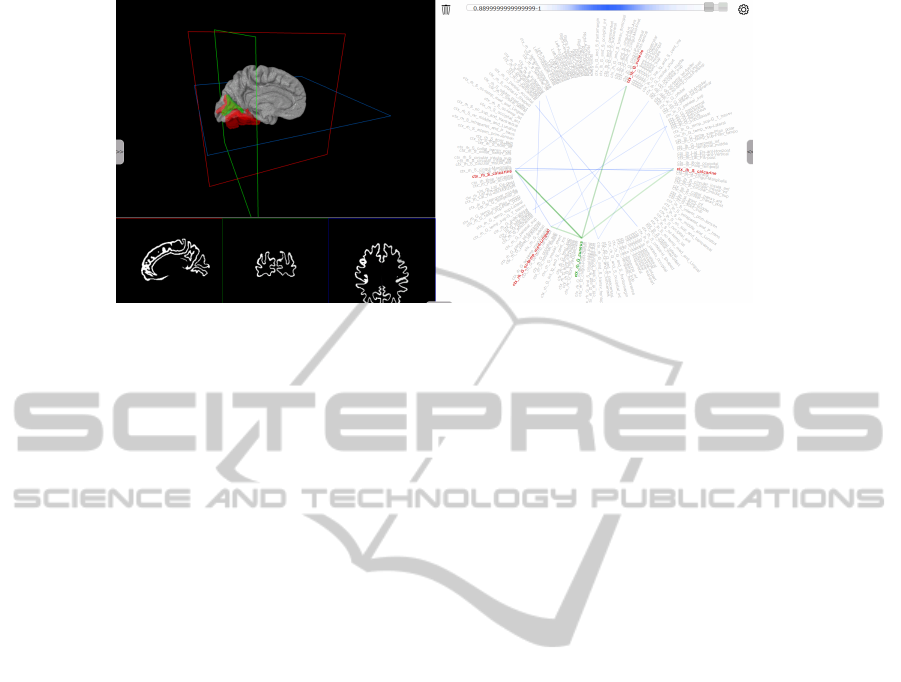
Figure 1: Overview of the visualization interface: Two representations are shown simultaneously, a 3D volume rendering and
a 2D visualization of the activity correlation, here a network visualization derived from a single correlation matrix. Sliders
can be used to define the range of correlation values that is modeled into the network. The subnetwork consisting of a selected
node (green) and its neighbors (red) is highlighted in the same color as the corresponding region in the 3D rendering, and the
axes in 3D view are positioned on the selected region.
measures, in particular when they express some value
that is aggregated over the whole graph, will not al-
ways adequately reflect the brain network character-
istics. However, even though networks with similar
topological properties may not be similar in terms of
neural activity, we still calculate these values as an
indicator to separate largely different networks.
Network properties that are calculated and con-
sidered useful for the analysis include the clustering
coefficient, the characteristic path length, the node
degree (weighted, unweighted, and distribution), the
number of isolated nodes, and centrality measures.
In addition, we take into account the anatomical
and functional annotations to calculate several values
for comparison of networks. We calculate an out-
lier fingerprint, based on the partition of the brain
into nine brain lobes derived from (Destrieux et al.,
2010). For each network we emphasize the amount to
which correlations per lobe differ significantly from
the whole network set. To this end, we first identify
the connections with a correlation that differs more
than the standard deviation from the median. For each
node, we sum up the number of such connections, and
identify the nodes for which that value again differs
more than the standard deviation from the respective
median number. As the nodes can be mapped to brain
lobes, we can aggregate the node values for each lobe
and thus create a fingerprint of length 9 that represents
the distribution of outlier nodes. The level of opac-
ity of a block in the fingerprint indicates the outlier
level, i.e. the stronger the color, the stronger the out-
lier characteristics regarding our measure. This fin-
gerprint then can be used to identify similar networks,
see Figure 4 right.
3.2 Visualization Components and
Interaction
Visualization Components. The main visualiza-
tion components for our approach are the 3D brain
rendering view and an abstract correlation data view,
which can be either a graph-based network view or
a matrix view, see Figures 1 and 2. Adding a net-
work visualization directly into the volume rendering
can make it difficult for the user to clearly perceive
the network structures. We conjecture that a side-by-
side visualization of network or matrix view and brain
anatomy will give the user a good overview and at
the same time allows for an intuitive interactive ex-
ploration of activity data. The assumption is based
on the idea that the 3D brain rendering allows intu-
itive orientation regarding the anatomical position and
functional annotation of the network nodes, while the
network can clearly depict the connection topology
and will thus help to detect patterns better than with
the 3D view alone. Hence our concept is based on
such a combination of a 3D brain rendering and a 2D
graph or matrix visualization, and we link the views
to enable the user to switch between views during in-
teractive exploration without a large cognitive effort.
In addition, our application features several inter-
active elements. These include pop-out panels for
the selection of networks and view details. Three
small subviews for the 3D rendering allow slice-based
browsing through the images, see Figure 1, bottom
left.
As part of our concept the precalculated network
properties for the currently selected network, or the
differences of these properties in comparison mode,
can be shown in a pop-out dashboard at the bottom of
IVAPP2015-InternationalConferenceonInformationVisualizationTheoryandApplications
134

Figure 2: Matrix view of the correlation data. The user can select correlations which then are highlighted both in the matrix
and the volume visualization. Red and blue color represent stronger correlation in one of the matrices under comparison.
Network statistics for a selected correlation threshold values are shown.
the screen. These values help to characterize a single
network, and can also be used for network compari-
son and classification.
We show a graphical representation of the outlier
fingerprint next to each network’s name in the net-
work list to enable the user to efficiently select most
relevant networks for analysis and comparison, see
Figure 4 right. Each brain lobe that contains outlier
nodes is depicted by a character that is colored ac-
cording to the number of outlier nodes. Blue color
represents nodes with a significant number of out-
lier nodes higher than the median, yellow represents
smaller numbers, and the intensity is varied depend-
ing on the number.
Interaction and Navigation. In the exploratory
workflow, a user can freely select networks for vi-
sualization and comparison. A correlation value in-
terval can be chosen by using the slider element lo-
cated on top of the network view, allowing the user
to define the range of the values used to model the
current network. Whenever the user adjusts the slider
setting, the corresponding network is displayed in the
network view. To support the user in choosing a use-
ful threshold, the distribution of correlation values is
mapped onto the slider by means of a color shading.
In comparison mode, two shadings show the different
distributions in the networks on the slider, see Fig. 3.
All views are coordinated and linked by a global
selection mechanism. In the 2D and 3D views, users
can select regions, either represented as a node in the
2D view, or as a rendered region in the 3D view. Se-
lecting elements in the visualization facilitates focus
on substructures of the network, e.g. the neighbor-
hood of a node in the network, or the subnetwork
corresponding to a brain region. Selection triggers a
highlighting by a change in colors for the selected re-
gion and its neighbors in the current correlation net-
work in the 2D and 3D views.
The visual cues to highlight elements during the
interactive exploration process are kept consistent
over all views. The selected node together with its lo-
cal neighborhood are emphasized in the network view
by a change in the color, see Figure 1, and the corre-
sponding brain regions are colored in the same way.
All colors are user-adjustable.
The volume rendering allows the user to freely
navigate within the 3D visualization that shows the
brain’s anatomy, rotating and zooming to the region
of interest (ROI). The volume axes in the 3D view are
automatically positioned to indicate the selected fo-
cus region, thereby also updating the slice views to
show the current selection. By interacting with the
slice subviews, the user can move the volume axes
and scroll through all individual slices. An ROI at-
las can be mapped onto the 3D rendering that allows
links back from selected brain regions to the network
visualization.
By changing the setting in the pop-out panels,
the user can select different networks for visualiza-
tion and comparison, and also toggle the visibility
of the volume rendering of white matter and pial
for each hemisphere depending on the user’s require-
ments. Both the network and the matrix view sup-
port a comparison mode that allows visualization dif-
ferences between selected networks. A color coding
scheme is used to show differences and similarities,
see Fig. 3 for the network view. For the compari-
son network described in Section 3.1, the edges with
a difference value within the threshold are drawn in
red and blue, where the color is determined by which
network has the higher correlation value. Strong cor-
relations occurring in both networks are drawn in yel-
CereVA-VisualAnalysisofFunctionalBrainConnectivity
135
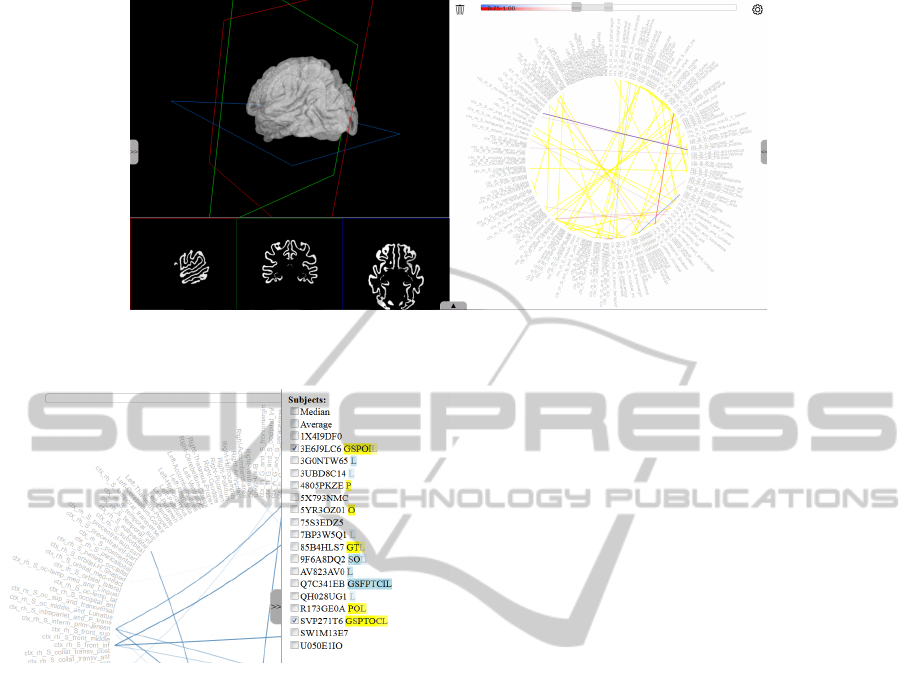
Figure 3: Network comparison combining similarities and dissimilarities for two networks. Differences above the user-defined
threshold value are shown in blue and red, common patterns in yellow.
Figure 4: Using the outlier fingerprint to analyze correla-
tion data. The fingerprint shows significant differences and
similarity both of a network to aggregated networks and be-
tween several pairs of individual networks, e.g. the net-
works labeled 3E6J9LC6 and SVP271T6 (selected for vi-
sual comparison) are quite similar regarding the outliers,
whereas SVP271T6 and Q7C341EB differ largely.
low. This helps the user to distinguish between net-
works that are similar besides a few differences and
networks that vary a large amount. The opacity of
the graphical edge representation depends on the dif-
ference values, such that more prominent patterns are
emphasized. This way, differences and their ampli-
tude are highlighted in the 2D view, allowing refined
filtering or selection of ROIs.
Network Layout. CereVA uses a circular layout for
graph visualization as the default layout, as it is estab-
lished in biomedical network visualization, including
recent iterations of brain network visualization tools
(Margulies et al., 2013). The view allows relatively
compact visualizations with a static aspect ratio, facil-
itating the side-by-side presentation. In order to min-
imize the effort to switch between 2D and 3D view,
we use a fixed node order that reflects the anatomical
location — clockwise from the top it shows left hemi-
sphere, right hemisphere, then central regions. The
membership and vicinity of network nodes with re-
spect to the brain regions are shown in the 3D view.
For an improved visualization of the connection pat-
terns, we apply edge bundling, where the tension dy-
namically adjusts for the number of visible edges, to
visually aggregate edges with a similar route. While
the single edge representations still exist in the visu-
alization, the bundling allows better identification of
global connectivity patterns by joining similar edge
sections to bundles, while also reducing visual clut-
ter.
We assume that for an in-depth visual analy-
sis of more complex patterns, different layout meth-
ods might prove more valuable, and are therefore
investigating the use of other methods for filtered
subnetworks. As such layouts might be subject to
constraints, constraint-based methods of the Web-
CoLa (Webcola, 2014) library might be best suited
to provide flexible layouts including data-driven con-
straints, e.g. to emphasize a spatial clustering of the
brain regions in the network layout.
3.3 Implementation
Our implementation allows the user to visualize rest-
ing state networks in the context of the brain’s
anatomy, and to compare networks using a node-link
representation. In addition, the raw correlation data
can be visualized and compared in a matrix view. The
comparison can be done visually in both the node-link
and the matrix view, while data- and network-based
indicators highlight network similarities and differ-
ences, facilitating analysis tasks A and B.
We created CereVA as a web-based solution, as
IVAPP2015-InternationalConferenceonInformationVisualizationTheoryandApplications
136

it can be easily made available and updated, without
the need to download and install an application. As a
web-based solution, we expect it to have advantages
when collaborative work on a data set is required, and
when interactive analysis of larger data sets can be
supported by cloud-based computing.
Our implementation uses several libraries for the
different parts of our interface. For the rendering of
3D visualizations in the browser we employed We-
bGL (WebGL, 2013). WebGL enables the browser di-
rect access the Graphics Processung Unit (GPU), re-
sulting in more seamless interaction and performance
compared to non-GPU implementations. The XTK li-
brary (XTK, 2014), a wrapper for WebGL, is used as
it is specifically designed to support rendering of 3D
medical image data. For the graph and matrix visu-
alization and layout we used the implementation pro-
vided by D3(D3, 2014), a well-established library for
web-based graph drawing.
4 DISCUSSION
To derive meaningful information from network visu-
alizations, the networks need to be modeled in an in-
tuitive way that facilitates knowledge discovery. Our
preliminary implementation allows the user to select
an interval for the correlation values that is used to
filter a subnetwork of the data set. While we think
that it is useful to allow the user such a filtering in
an interactive fashion to focus on ROIs, the search for
a reasonable threshold should be guided by an auto-
mated analysis. We use a constant threshold to start
with, and give the user visual hints based on network
characteristics to support their search.
Network comparison is an active area of research,
and there is no obvious optimal visualization of brain
activity networks. While our solution presents the dif-
ference and similarities in a single cumulative graph-
based view, separate views, for example individ-
ual graphs representing specific characteristics (small
multiple graphs), might be useful to quickly detect
patterns and to identify outliers. We aim to inves-
tigate further whether calculations such as weighted
maximum common subgraphs help to better identify
common patterns. The reason for this suggestion is
that we could see that for several pairs of threshold-
filtered networks, the dominant differences consisted
only of one or two additional edges. Our network
analysis so far does not make use of the fact that we
have labeled graphs, i.e. we have some specific brain
location associated with each node of the network. As
the combination of this information may prove useful,
we will include it in the analysis in future steps.
Aggregation, including of subnetworks, may re-
duce the complexity and emphasize patterns in the
poplation. However we have not seen enough evi-
dence so far of which network characteristics could be
subject to aggregation. We aim to answer this ques-
tion in ongoing discussions with collaborating neu-
roscientists. This might also help to further support
analysis task C, in particular when clustering and clas-
sification methods will be included, as this task is so
far mainly supported by giving the user information
on network and correlation characteristics. Finally
larger neighborhoods or subnetwork patterns than the
distance one neighborhood might be of interest for
highlighting or semi-automated selection.
5 OUTLOOK
In this paper, we presented CereVA, a preliminary
concept for the visual analysis of brain activity corre-
lation data. CereVA is implemented as a web service
that provides an interactive visual interface for data
exploration. Our approach currently supports a ba-
sic correlation matrix format as input, and we plan to
provide import and export of data and visualizations
in standard formats, e.g as provided by the large brain
research projects. Further, we aim to integrate addi-
tional knowledge that is available for example in such
public databases or publications.
In order to extend our solution towards a visual
analysis approach, we will embed additional infor-
mation for analysis and visualization. This will in-
clude an analysis of time series data, as we believe
that an analysis will provide further insight into the
variation of correlations for a single subject, and thus
allow experts to derive better characterizations of sim-
ilarities and differences across the population. Visu-
alizations and interactions based on a decomposition
of the network, e.g. using modularity clustering or
functional aggregation, will be considered for addi-
tion to the system. While the linked complementary
2D and 3D views demonstrated its capabilities to pro-
vide a 3D anatomical reference for the visualization
of activity networks, adding information from struc-
tural MRI and diffusion tensor imaging might greatly
improve the analysis of individual brain activity, e.g.
after brain damage.
To assess the usefulness of our system beyond the
feedback from domain experts during the develop-
ment, and to make decisions on the further implemen-
tation and features, we will perform a formal evalu-
tion with domain experts. This paper presented our
findings from the use of resting-state MRI networks,
however our system is not limited to this data type and
CereVA-VisualAnalysisofFunctionalBrainConnectivity
137

we will explore extension of our system for use with
different types of networks.
REFERENCES
Alper, B., Bach, B., Riche, N. H., Isenberg, T., and Fekete,
J.-D. (2013). Weighted graph comparison techniques
for brain connectivity analysis. In Mackay, W. E.,
Brewster, S. A., and Bødker, S., editors, CHI, pages
483–492. ACM.
B
¨
ottger, J., Schurade, R., Jakobsen, E., Schaefer, A., and
Margulies, D. S. (2014). Connexel visualization: a
software implementation of glyphs and edge-bundling
for dense connectivity data using braingl. Front Neu-
rosci, 8(15).
BSA (2014). Brainspan Atlas. See
http://www.brainspan.org/.
Bullmore, E. and Sporns, O. (2009). Complex brain net-
works: graph theoretical analysis of structural and
functional systems. Nat. Rev. Neurosci., 10(3):186–
198.
Chang, H.-M., Chiang, A.-S., Didimo, W., Lin, C.-Y., Li-
otta, G., and Montecchiani, F. (2013). On the robust-
ness of the drosophila neural network. In Network Sci-
ence Workshop (NSW), 2013 IEEE 2nd, pages 168–
171.
D3 (2014). Data Driven Documents. See http://d3js.org/.
Destrieux, C., Fischl, B., Dale, A., and Halgren, E. (2010).
Automatic parcellation of human cortical gyri and
sulci using standard anatomical nomenclature. Neu-
roImage, 53(1):1 – 15.
Greicius, M. D., Srivastava, G., Reiss, A. L., and Menon, V.
(2004). Default-mode network activity distinguishes
alzheimer’s disease from healthy aging: Evidence
from functional mri. Proceedings of the National
Academy of Sciences of the United States of America,
101(13):4637–4642.
Hagmann, P., Cammoun, L., Gigandet, X., Meuli, R.,
Honey, C. J., Wedeen, V. J., and Sporns, O. (2008).
Mapping the structural core of human cerebral cortex.
PLoS Biol, 6(7).
HBP (2014). The Human Brain Project. See
https://www.humanbrainproject.eu/.
Lee, H., Kang, H., Chung, M. K., Kim, B. N., and Lee, D. S.
(2012). Persistent brain network homology from the
perspective of dendrogram. IEEE Trans Med Imaging,
31(12):2267–2277.
Margulies, D. S., B
¨
ottger, J., Watanabe, A., and Gor-
golewski, K. J. (2013). Visualizing the human con-
nectome. NeuroImage, 80:445–461.
Nowke, C., Schmidt, M., van Albada, S., Eppler, J., Bakker,
R., Diesrnann, M., Hentschel, B., and Kuhlen, T.
(2013). Visnest – interactive analysis of neural ac-
tivity data. In Biological Data Visualization (BioVis),
2013 IEEE Symposium on, pages 65–72.
Onoda, K. and Yamaguchi, S. (2013). Small-worldness and
modularity of the resting-state functional brain net-
work decrease with aging. Neurosci. Lett., 556:104–
108.
Smith, S., Vidaurre, D., Beckmann, C., Glasser, M., Jenkin-
son, M., Miller, K., Nichols, T., Robinson, E., Salimi-
Khorshidi, G., Woolrich, M., Barch, D., Ugurbil, K.,
and DC, V. (2013). Functional connectomics from
resting-state fmri. Trends in Cognitive Sciences, 17.
Sorger, J., Buhler, K., Schulze, F., Liu, T., and Dickson, B.
(2013). neuromap – interactive graph-visualization of
the fruit fly’s neural circuit. In Biological Data Visu-
alization (BioVis), 2013 IEEE Symposium on, pages
73–80.
van den Heuvel, M. P. and Pol, H. E. H. (2010). Explor-
ing the brain network: A review on resting-state fmri
functional connectivity. European Neuropsychophar-
macology, 20(8):519 – 534.
Van Essen, D. C., Smith, S. M., Barch, D. M., Behrens,
T. E., Yacoub, E., and Ugurbil, K. (2013). The wu-
minn human connectome project: An overview. Neu-
roImage, 80(0):62 – 79. Mapping the Connectome.
Wang, J., Zuo, X., and He, Y. (2010). Graph-based network
analysis of resting-state functional mri. Frontiers in
Systems Neuroscience, 4(16).
Webcola (2014). WebCoLa. See
http://marvl.infotech.monash.edu/webcola/.
WebGL (2013). WebGL. See
https://www.khronos.org/webgl/.
XTK (2014). The X Toolkit. See
https://github.com/xtk/X#publications.
IVAPP2015-InternationalConferenceonInformationVisualizationTheoryandApplications
138
