
The Use of Extensible Markup Language (XML) to Analyse Medical
Full Text Repositories – An Example from Homeopathy
Thomas Ostermann, Marc Malik and Christa Raak
Institute of Integrative Medicine, Witten/Herdecke University, Gerhard-Kienle-Weg 4, Herdecke, Germany
Keywords: Extensible Markup Language, Homeopathy, Repertorisation, Software.
Abstract: Extensible Markup Language (XML) is one of the most popular web languages in the life science used for
for Semantic Data Analysis in various fields of clinical research. One of these fields is the processing of
medical full texts. To extract meaningful information out of natural texts is one of the challenges when
dealing with huge text repositories. We present an application of XML together with linguistic algorithms in
the processing of texts from a homeopathic materia medica. Our approach enables the user not only to
search within the symptom descriptions but also offers special features like sequential search within the
results or the comparison of homeopathic remedies. However user demands of day to day practice and terms
of information technology have both to be taken carefully into account to further develop this prototype.
1 INTRODUCTION
Extensible Markup Language (XML) is one of the
most popular semantic web language in the life
science with more than 900 publication between
1999 and 2010 in PubMed (Ostermann et al., 2014).
Today it is applied in knowledge transfer in the life
science (Murray-Rust, 2000) In this field XML has
managed to become an important tool in clinical
laboratory procedures (Saadawi and Harrison, 2003)
but its way into patient care still seems to be far
behind the possibilities XML is offering.
In particular the capability of Internet Browsers
to read, edit and analyse XML documents creates a
variety of opportunities for Semantic Data Analysis
(SDA) facilities to be incorporated into clinical
applications (Bompani et al., 2002).
XML, when combined with web services,
semantic data analsis and scripting languages such
as Java script, can be used to offer a huge amount of
functionality for the user including text retrieval and
the generation of summary data through a standard
web-browser.
With regards to medical full texts, searching for
a certain information sometimes is crucial. As
already pointed out by Grivell in 2002 “natural
language provides a considerable challenge for
algorithms to extract meaningful information from
natural text.”
This even more becomes a complex problem
when dealing with huge repositories from the field
of traditional medical systems. In particular,
machine readable dictionaries with a codification of
domain knowledge and literature metadata in
accordance with a generic and extendible XML
scheme model have been shown to be suitable in this
context Ostermann et al., 2009). One open problem
in in this context is the semantic processing of the so
called Materia Medicae. Such repositories contain
structured data on medical symptoms and the
corresponding remedies i.e. from the field of
phytotherapy or, like in our case, from homeopathy.
With a tradition of 200 years of patient care,
homeopathy is one of the oldest integrative medical
systems in the field of Traditional European
Medicine. An essential part of homeopathic case
taking is the conduction of a comprehensive
anamnesis followed by individualized finding of a
remedy that fits the conditions the patient describes.
This is called repertorisation and today is done with
the help of computer programs using modern
database technology (Ostermann et al., 2012).
Accoring to our own review and with respect to
other personalized approaches in e-health (Lee et al.,
2008), XML-based processing of such vast resources
might be beneficial in the complex process of
homeopathic prescribing.
219
Ostermann T., Malik M. and Raak C..
The Use of Extensible Markup Language (XML) to Analyse Medical Full Text Repositories – An Example from Homeopathy.
DOI: 10.5220/0005484002190224
In Proceedings of 4th International Conference on Data Management Technologies and Applications (DATA-2015), pages 219-224
ISBN: 978-989-758-103-8
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)
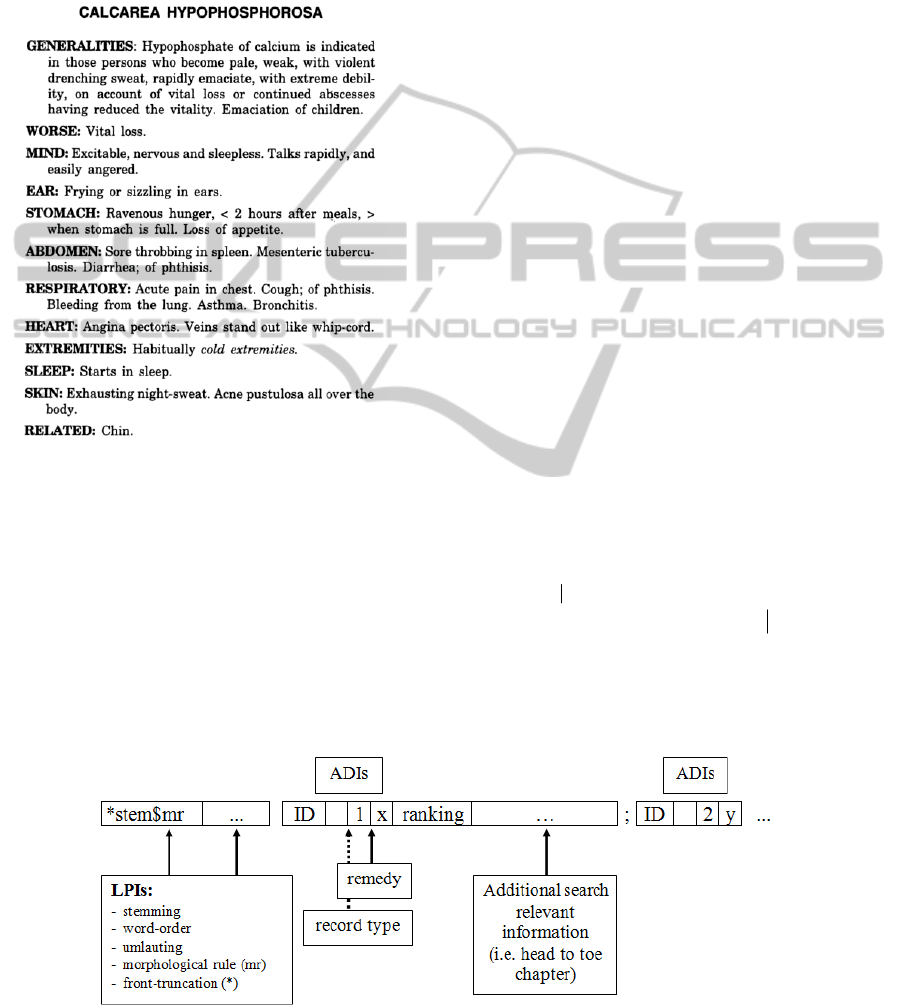
2 MATERIAL AND METHODS
Phataks’ Materia Medica contains 419 different
homeopathic remedies described in their symptoms
in a Head-To-Toe Scheme. Fig. 1 illustrates this
structure on the example of Calcerea
Hypophosphorosa (CaHPO
4
):
Figure 1: Description of the homeopathic remedy Calcerea
Hypophosphorosa in its symptoms in a Head-To-Toe
Scheme (Phatak 2011).
For every remedy a more or less detailed
description comparable to the one given in Fig. 1
was ripped into approx. 25.000 phrases resp.
sentences divided either by a full stop or a
semicolon. Phrases were processed into an index by
linguistic algorithms, and together with additional
discriminating information (ADIs) for example, the
logical subset (i.e. a remedy or a head to toe section)
a phrase belongs to, an inverted file structure is
created (Fig. 2; for an overview see Zobel and
Moffat, 2006).
In our example of Calcerea Hypophosphorosa
*stem$mr” denotes the decomposition of a search
term i.e. the search term “drenching” leads to
“drench$∼ing”. Note that front truncation
information (marked with a “*”) and umlauting are
features specially designed for German language and
do not occur quite often in English language (i.e. for
composition terms like “Night-Sweat” which in the
case of “nightsweating” would become
*sweat$∼ing).
Another algorithm decomposes phrases into their
linguistic and grammatical entities. In the example
of figure one the term “Acute pain in chest”, the
word “pain” is the subject modified by “in chest”
and restricted by “acute”, whereas in the phrase
“chest pain in acute bronchitis” constructed by the
same words, the roles of modifying and restricting
terms change. Thus, although the textual phrase
consists of the same words, the ranking of the search
results will be different because of the grammatical
relation between the words. Words therefore are no
longer regarded as textual 'singularities', but are
recognized in their syntactic-semantic interrelation.
(Zillmann, 2000).
The complete vocabulary of the material medica
with exclusion of content notes such as page
numbers and stopwords such as "the",”an” or "and",
which were filtered out prior to the processing is
stored this way in a Prefix-B-tree which stores the
strings in lexicographic order with head and tail
compression as described above (Bawa et al., 2005).
Let
{}
DddC 1==
denote the corpus containing a
total of D datasets d and let
{
}
nidwwW
d
i
d
id
1; =∈=
denote the set of all words
d
i
w
of the dataset. Then an
address vector
()
k
d
i
d
i
aaaww
,,
2,1
=
is defined where
k
aaa ,
2,1
denote additional coded information about
syntactical and morphological attributes of the word.
Figure 2: Schematic description of an inverted file structure with linguistic processing information in the front and
additional discriminant information (ADIs) in the back of the coding (adopted from Ostermann et al., 2009).
DATA2015-4thInternationalConferenceonDataManagementTechnologiesandApplications
220

Then
{
}
niwW
d
jd
~
1;
~
==
is called the index of the
dataset d and
d
WW
~
~
=
is called the index of the
whole database. Finally the terms
{
}
mjssS
jj
1; ==
of
the search query S are defined. Again, according to
the construction of
d
i
w
a search vector
(
)
kjj
aaass
,,
2,1
=
is defined.
The quality function is then defined as
()
()
==
≤+=
m
j
n
i
j
d
iimd
swqQSQ
11
100,
,
where
(
)
j
d
ii
swq
,
denote additional quality criteria
functions (i.e. correct flexion, correct position and
order of the words, containment in a compound
word, irregular plurals) and
m
Q
denotes a fixed
accuracy parameter (in our case Q
m
=45). Together
with a quality threshold
0; >−=
δ
δ
mmt
QQ
defined
as the lower bound for the quality of a dataset, every
dataset d with a value
()
td
QSQ ≥
is presented as a
result of the search query S. Datasets with a higher
ranking then will be placed in a higher position than
those with a lower ranking (Fig. 3).
One important feature is given by the “Search
within the results” feature. As homeopathic
diagnosis is based on a sequential process of adding
up symptoms of a patient, this process has to be
integrated in the search architecture. Thus the
process described in Fig. 3 is repeated within the
search results: In a first step the therapist submits a
query comprising terms describing the patients
symptoms i.e. “throbbing headache in the morning”.
This query is then processed via linguistic
algorithms resulting in a list of possible remedies.
The therapist now can modify his search query by
adding a second symptom i.e. “chest pain with
cought”. Based on the results of the first search
query given by the record-IDs, the search engine
runs a second query which refines the first query.
The process is repeated until a lower bound of 10
remedies is reached.
The results of a search query are represented in
XML-structured metadata and delivered to the
clients web browser. At the client side XML-data is
processed via Extensible Stylesheet Language
Transformations (XSLT). Fig. 4 displays a result for
the search query “starker Nachtschweiß” (engl:
exhausting night-sweat”).
As can be seen in this example, Remedies are
ranked higher, when the search query “starker
Nachtschweiß” is not modified by other descriptors.
This is the case in the first three hits “Taraxacum”,
“Ledum palustre” and “Ammonium muriaticum”
which completely contained the phrase “starker
Nachtschweiß” (or its plural) seperated by
punctuation from other phrases and thus Q = 100%.
In the next four hits the phrase “starker
Nachtschweiß” is modified i.e. in hit 7: “starker
Nachtschweiß, nach Schwefel riechend” (engl:
smelling like sulfur). Thus the quality function is
reduced to Q=98% resp. 97%. In the last case of
“Calcera phosphorica” Q is reduced to 88% because
in the full text the restricting adjective “exhausting”
is parenthesized: [starker].
Figure 3: Schematic description of the relationship between inverted file and document quality scores (adopted from Zobel
and Moffat 2006).
TheUseofExtensibleMarkupLanguage(XML)toAnalyseMedicalFullTextRepositories-AnExamplefrom
Homeopathy
221

Figure 4: Representation of the search results for “starker
Nachtschweiß” (engl.: “exhausting night-sweat”. Please
note that our example “Calcerea Hypophosphorosa” is
known in the German Version of Phatak’s material medica
as “Calcarea phosphorica”.
Another example is given by the search query
“klopfende Kopfschmerzen” (engl: “throbbing
headache”) in Fig. 6.
Again the first hit finds the complete search
phrase “klopfende Kopfschmerzen” as a standalone
phrase and thus Q=100%. In hit number two the
search phrase is modified by “nach der Menses”
(engl: “after menses”) resulting in a quality function
value of Q= 97%. In hit three “Crocus sativus” the
search term „klopfender Kopfschmerz” is found
completely, however it is restricted by “pochender”
(engl. “beating”). This restriction reduces Q to 93%.
In the results 4-6 we have the same formal
restriction like in hit three, however the search
phrase is additionally modified by “nervös,…”
(engl.: “nervous”) in hit four “Melilotus”,
“schlimmer…”( engl.: “worsened”) in hit five
“Ledum palustre” and “besser…” (engl.:
“improved”) in hit six “Pyrogenium”. In hit 7
“Lycopodium” we have a German synonym
“Klopfendes Kopfweh” which later in the sentence
is explained as “Kopfschmerzen”, the exact wording
of the search phrase. Hit number eight “Calcera
carbonica“ quite intuitively shows that “klopfende
Kopfschmerzen” also finds „Kopfschmerzen tief im
Gehirn, klopfend“ (engl. “headache, throbbing deep
in the brain”) leading to a further reduction of the
quality function Q to 88%. Finally but is even more
important the search term “klopfende
Kopfschmerzen” also finds the composition term
“klopfender Stirnkopfschmerz” (engl. “throbbing
frontal headache”) of Lac defloratum (Hit 9)
although with a lower ranking of Q=84% (See
Figure 6 for the search results).
<record>
<RecordRelevance>91</RecordRelevance>
<RecordNumber>5</RecordNumber>
<RecordLink>/cgi-
bin/CiXbase/phatak5/CiXbase_search?act=search
8;search=sqn&sqn=00021005</RecordLink>
<RecordID>00021005</RecordID>
<DataType>c</DataType>
<RecordType>o</RecordType>
<TitleID>00020968</TitleID>
<cTitle>Ledum palustre</cTitle>
<cKopf>Kopf</cKopf>
<cKeyword>Wütender, klopfender Kopfschmerz,
schlimmer durch die geringste
Kopfbedeckung</cKeyword>
</record>
Figure 5: Extract of the XML-representation of the search
result no. 5 for “klopfende Kopfschmerzen”.
Figure 6: Representation of the search results for
“klopfende Kopfschmerzen” (engl.: “throbbing headache”.
DATA2015-4thInternationalConferenceonDataManagementTechnologiesandApplications
222

Figures 5 and 6 also demonstrate that structure
and layout are separated. This leads to an almost
complete interoperability of the metadata and eases
the transfer to other information systems like mobile
applications.
3 RESULTS
Based on the results of published homeopathic cases
we were able to reproduce the results of
repertorisation with the E-Phatak. In ten cases listed
in Table 1 we were able to reproduce the wanted
results with our prototype. A comparison with
conventional repertory software “RADAR:
easyRep” and the electronic version of the
“Bönninghausen’s Therapeutic pocketbook” also
found promising results
A first evaluation by a focus group of
homeopathic physicians and healing practitioners
moreover revealed that all evaluators found
sequential search to be the key feature and the
innovative element of the E-Phatak which should be
the subject of further investigations and
implementations.
Table 1: Comparison of ranking results of the E-Phatak
compared with easyRep and the electronic version of the
“Therapeutic pocketbook”
On the other hand ligustic ranking was found to
be difficult to understand for the therapist using the
phatak in daily patient care.
One major issue was claimed from the focus
group to be the most crucial point when working
with the E-Phatak: searching for “throbbing
headache” might i.e. miss synonymous phrases like
“hammering headache” or “knocking headache”,
which already has been discussed in the field of full
text searching from Beall (2008).
In our case of figure 6 this can be seen in hit
number 7 “Lycopodium”. Although from a
therapeutical point of view it contains what the
therapist was looking for (namely “throbbing
headache”), it misses a higher ranking due to the
linguistic processing, which does recognize a
similarity in the semantics but not in the meaning of
the phrase itself.
4 CONCLUSIONS
Information technology nowadays has reached
almost every part of patient care. In particular
electronic systems for decision support of physicians
and therapists are major issues in health care
informatics. However a recent review of Romano
and Stafford (2011) on the impact of such systems
on national ambulatory care quality suggests “no
consistent association” between the use of clinical
decision support systems and better quality in patient
care if they are used isolated and are not integrated
in routine daily care.
In the field of homeopathy, electronic decision
support systems have been introduced and
incorporated quite early in patient care with first
affordable software applications on the market in the
early 1990
th
. Moreover homeopathy has always
made use of the most innovative technology
available (Ostermann et al., 2012).
We were able to show how full text searching in
homeopathic text repositories can be achieved using
XML and XSLT. This technical realisation enables
the user not only to search within the symptom
descriptions but also offers special features like
sequential search within the results or the
comparison of homeopathic remedies.
However user demands of day to day practice
and terms of information technology have both to be
taken carefully into account to further develop this
prototype. In particular with regards to several
systematic reviews (Boulos et al., 2011, Free et al.,
2013), mobile health applications should be taken
into consideration for a second launch of the E-
Phatak.
ACKNOWLEDGEMENTS
We would like to thank Elsevier-Publishers,
Germany and the Software AG-Foundation for their
support of the project.
TheUseofExtensibleMarkupLanguage(XML)toAnalyseMedicalFullTextRepositories-AnExamplefrom
Homeopathy
223

REFERENCES
Bawa M, Condie T, Ganesan P. LSH forest: self-tuning
indexes for similarity search. Proceedings of the 14th
international conference on World Wide Web 2005;
651-660.
Beall J. The weaknesses of full-text searching. The Journal
of Academic Librarianship 2008, 34(5), 438-444.
Bompani L., Ciancarini P., Vitali F. XML-based
Hypertext Functionalities for Software Engineering.
Annals of Software Engineering 2002, 13:231–248.
Boulos, MN, Wheeler S, Tavares C, Jones R. How
smartphones are changing the face of mobile and
participatory healthcare: an overview, with example
from eCAALYX. Biomedical engineering online
2011, 10(1), 24.
Free C, Phillips G, Watson L, Galli L, Felix L, Edwards
P., Haines A. The effectiveness of mobile-health
technologies to improve health care service delivery
processes: a systematic review and meta-analysis.
PLoS medicine 2013, 10(1), e1001363.
Grivell L. Mining the bibliome: searching for a needle in a
haystack? EMBO reports 3.3 (2002): 200-203.
Halpin H, Thompson HS. One document to bind them:
Combining xml, web services, and the semantic web.
Proceedings of the 15th international conference on
World Wide Web; 2006; 679-686.
Lee CO, Lee M, Han D, Jung S, Cho J. A framework for
personalized Healthcare Service Recommendation.
Proceedings of the HealthCom 2008. 10th
International Conference on e-health Networking,
Applications and Services 2008; 90-95).
Murray-Rust P, Rzepa HS, Wright M, Zara S. A universal
approach to web-based chemistry using XML and
CML. Chemical Communications 2000; 16: 1471-
1472.
Ostermann T, Raak CK, Matthiessen PF, Büssing A,
Zillmann H. Linguistic processing and classification of
semi structured bibliographic data on complementary
medicine. Cancer Inform. 2009 Jul 6;7:159-69.
Ostermann T, Raak C, Malik M. Software technology
applications for repertorisation in homeopathy–a
systematic review. European Journal of Integrative
Medicine 2012; 4: 197.
Ostermann T, Raak C, Malik M. Application of extensible
markup language (XML) in medical research: A
bibliometrical analysis. Proceedings of the 7th
International Conference on Health Informatics,
HEALTHINF 2014; 478-483.
Phatak SR. Materia Medica of Homoeopathic Medicine,
Second Revised & Enlarged Edition. Jain Publishers
2011.
Romano MJ, & Stafford RS. Electronic health records and
clinical decision support systems: impact on national
ambulatory care quality. Archives of internal medicine
2011, 171(10), 897-903.
Saadawi G, Harrison JH Jr. XML syntax for clinical
laboratory procedure manuals. AMIA Annu Symp
Proc. 2003:993.
Zillmann, H. 2000. Information Retrieval and Search
Engines in Full-Text-Databases. Liber Quarterely, 10:
335-41.
Zobel J and Moffat A. Inverted files for text search
engines. ACM Comput. Surv., 38(2), 2006.
DATA2015-4thInternationalConferenceonDataManagementTechnologiesandApplications
224
