Investigation into Mutation Operators for Microbial Genetic Algorithm
Samreen Umer
University College Cork, Western Road, Cork, Ireland
Keywords:
Microbial Genetic Algorithm, Evolutionary Algorithms, Mutation.
Abstract:
Microbial Genetic Algorithm (MGA) is a simple variant of genetic algorithm and is inspired by bacterial con-
jugation for evolution. In this paper we have discussed and analyzed variants of this less exploited algorithm
on known benchmark testing functions to suggest a suitable choice of mutation operator. We also proposed a
simple adaptive scheme to adjust the impact of mutation according to the diversity in population in a cost ef-
fective way. Our investigation suggests that a clever choice of mutation operator can enhance the performance
of basic MGA significantly.
1 INTRODUCTION
The idea of using evolutionary principles for auto-
mated problems was first originated in the middle of
20th century. Dr. L.J Fogel, known as father of evo-
lutionary programming, presented first evolutionary
technique in his dissertation (Fogel et al., 1966). Later
on, this area got significant attention and different di-
alects of this technology were evolved. Since then
swarm based optimization and nature inspired algo-
rithms marked their significance in computational sci-
ences. In recent decades these techniques have gain
much popularity in terms of optimization applica-
tions, system design and scheduling operations. Ge-
netic algorithms are evolutionary computation tech-
niques which mimics the evolution process to gener-
ate useful solutions for optimization problems (John,
1992). Many variants for genetic algorithms have
been introduced to satisfy different constraints ac-
cording to different situations. Here we will discuss
about one of the variants of genetic algorithm called
Microbial Genetic Algorithm (MGA).
Harvey introduced the idea of bacterial recom-
bination or infection as a substitute to inheritance
from parents in genetic algorithm (McCarthy, 2007)
and called it as microbial genetic algorithm (Harvey,
2011). Motivation behind this variant was to produce
a minimalist algorithm still containing all the char-
acteristics of a true genetic algorithm. Since then,
bacterial or microbial evolution based algorithms
have been used successfully by many researchers and
scientists. However any investigation on MGA’s per-
formance or its operators is not available in literature.
We conducted this study to verify the behaviour of
basic MGA and MGA with newer mutation operators
to solve unconstrained optimization problems. We
introduced a simple adaptive mutation scheme and
results conclude that a clever selection of MGA
operators can improve its performance notably.
2 MICROBIAL GENETIC
ALGORITHM
MGA is a simpler variant of GA. Main difference
between these two is different recombination opera-
tor. In MGA, initially parents are selected from gen-
erated population using a selection scheme and, are
then evaluated on the basis of their fitness to mark a
loser and winner. Unlike basic GA, MGA crossover
genes in such a way that the winner is left intact and
some genes are transferred to the loser with a defined
crossover probability. In this way the elitism is main-
tained and assumingly good genes are transferred to
the next generation. Mutation is then carried out on
infected loser to keep the diversity in the population.
The whole routine can be repeated until the desired
results are achievedor up to maximum number of iter-
ations allowed. Figure 2.1 illustrates one tournament
or life cycle of MGA.
2.1 Generating Population
Like in any other evolutionary algorithm, the first step
is to generate a potential population within domain
Umer, S..
Investigation into Mutation Operators for Microbial Genetic Algorithm.
In Proceedings of the 7th International Joint Conference on Computational Intelligence (IJCCI 2015) - Volume 1: ECTA, pages 299-305
ISBN: 978-989-758-157-1
Copyright
c
2015 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
299

Figure 1: One MGA Cycle.
of the problem. Size and diversity of the popula-
tion plays a crucial role in the performance of any
algorithm. Larger the population ensures greater di-
versity but more computations.Small population can
converge in less time but can also lead to premature
convergence. Many studies have been conducted to
generalize an optimal population size but mostly it
depends on the dimension of the problem and number
of maximum iterations allowed. Generally the popu-
lation is generated from a uniform distribution within
prescribed range and is unbiased.
2.2 Selecting Parents
A variety of selection technique is available for ge-
netic algorithms. Some possibilities are stochas-
tic universal sampling (Baker, 1985), Ranked selec-
tion (Whitley et al., 1989), Roulette wheel selection,
and Truncation selection (Goldberg and Deb, 1991).
However we have used ranked roulette selection as
our selection scheme in experiments.
2.2.1 Ranked Roulette Selection
Roulette scheme of selection is one of the commonly
used scheme among the researchers. In this method,
the probability of selection P
i
is assigned to the mem-
ber i of population size n in accordance to their fitness
value f
i
such that
P
i
=
f
i
∑
n
j=1
f
j
(1)
For finding optimal minimums this formula can be
manipulated as
P
i
=
∑
f − f
i
∑
f
(2)
But in case of non positive values unfortunately
this equation can not work leaving us with the im-
plication of some kind of scaling or scoring of fit-
ness values. One possible solution to such problem
was proposed by (Al Jadaan and Rao, 2008) as an
improved selection operator. The population is first
evaluated and sorted in accordance to the rank based
on fitness value and then the probabilities are assigned
using following formula
P
i
=
2R
i
Pop(Pop+ 1)
(3)
Where P
i
is the probability of individual i with
rank R
i
from population with with population size
Pop for being selected as a parent. A simple ranking
scheme can be where rank of a member is its position
in sorted population according to fitness.
The above scheme is used to select parents for
each cycle or tournament. Parents are then evaluated
and assigned as winner and loser for their survivabil-
ity in next generation.
2.3 Recombination
In each cycle the population is replaced with new gen-
eration. The selection scheme mentioned above gives
the parents to breed for children who will be intro-
duced in population to make new generation. In con-
ventional GA this has been the normal way of each
life/death cycle where the parents are replaced with
the children and thus genes are transferred vertically
down to the next generation. However in Microbes,
breeding is not in similar fashion. The organisms or
microbes reproduce by binary fission and further ex-
change their genetic material via a process called bac-
terial conjugation. What happens in this phenomenon
is that the fittest bacteria most commonly the one who
has developed resistance to the antibiotics makes a
contact with the weaker vulnerable bacteria and trans-
fer its resistance in terms of plasmid to it. MGA
uses this concept of parallel gene transfer to evolve
instead of horizontal gene transfer in conventional
GA. The fittest among the parents transfer its genes
to the weaker thus the new chromosome is actually
the weaker parent with some of its genes replaced by
genes from the winner.
2.4 Mutation
Mutation is responsible for maintaining the diver-
sity in population and preventing premature conver-
gence. Extensive research and studies have been done
already to provide different mutation operators and
proved to be useful in most cases. Following oper-
ators are reviewed here for further use in experimen-
tation ahead.
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
300
2.4.1 Uniform One Point Mutation
One random gene from the member which in this case
is the infected loser, is replaced with a new gene. In
case of binary MGA the gene is a bit so mutation
means just flipping the bit. However in case of real
number or integers, a new gene is generated from the
possible range randomly. Uniform one point mutation
is a technique in which at one random locus, the gene
is replaced by a new one generated randomly from a
uniform distribution.
2.4.2 Gaussian One Point Mutation
Instead of uniform distribution other distributions like
Gaussian can be used for generating the new gene. A
classical Gaussian mutation operator was developed
by Rechenberg and Schwefel in which a scaled Gaus-
sian normally distributed number is added to the pre-
vious value of gene (Back and Schwefel, 1993) . This
method requires information about mean and vari-
ance of desired distributionto produce a random gene.
Generally value of mean is set to be 0 with variance of
0.2 to 0.8. the formula for Gaussian density function
with mean µ = 0 and δ variance is given by
f(x) =
1
√
2πδ
2
e
−x
2
2δ
2
(4)
2.4.3 Cauchy One Point Mutation
For some functions with relative minima that are far
apart the small mutations of uniform or Gaussian dis-
tributions may lead to premature convergence to local
minima. For this reason (Yao et al., 1999) devised
an alternative to the Gaussian mutation by using a
Cauchy distributed random number instead. Cauchy
distribution with mean µ = 0 is defined as
f(x) =
t
π(t
2
+ x
2
)
(5)
Where t is the scaling factor and usually set as 1.
This allows for larger mutation and can help prevent
premature convergence and search the search space
faster. However there is less probability of smaller
mutations in the neighbourhood of the parent leading
to a less accurate local search.
2.4.4 Forced Adaptive Guassian Mutation
Experiments suggest that MGA converges faster in
earlier stages but with time as population tends to be
more similar, the same mutation operator does not re-
main as effective. Many researchers have proposed
adaptive or forced mutation operators for enhancing
its over all performance. In (Hatwagner and Horvath,
2012) a new forced mutation operator has been intro-
duced for bacterial evolutionary algorithm based on
relationship between diversity of population and vari-
ance of mutation operator. Howevercalculating diver-
sity in each iteration incurs additional computational
overhead and delays. Inspired by their technique, we
designed a mutation operator where we set thresholds
to calculate diversity in the population and adjust mu-
tation accordingly. The diversity of population is cal-
culated using formula given in (Miller and Goldberg,
1995).
d
i, j
=
s
g
∑
k=1
(
x
i,k
−x
j,k
X
k,max
−X
k,min
) (6)
D =
1
Pop −1
Pop
∑
i=1
d
i,best
(7)
Any mutation scheme can be coupled with this
technique however we used gaussian scheme where
the variance is controlling parameter of mutation. Ob-
jective for designing this operator is to improve the
diversity D in earlier evaluations and tune to fine po-
sition in the later stages. So when the population is
generated it usually has higher diversity at that point
we can directly relate the variance σ with D allowing
MGA routine to get variety in its early generations.
With higher variance we observed no improvement in
this situation where as low variance in such condition
showed better refine. Using following formula where
Variance σ increases with diversity up to a threshold
value lets say D = 0.5 and then start decreasing lin-
early .
σ = α.min(D, 1−D) (8)
α is the scaling constant with values 0 ≤ alpha ≤ 1
. σ obtained is then used in equation (4) to facilitate
gaussian mutation.
2.5 Replacing Previous Generation with
New One
Now comes the turn to introduce the infected and mu-
tated member into the population to proceed for next
cycle. In basic original GA, usually either both of the
selected parents are replaced with new crossed and
mutated offsprings or the worst of whole population
is replaced with the new offsprings. Howeverto main-
tain the elitism by keeping the winner intact and also
avoid losing all of the genes from loser, in basic MGA
loser chromosome is replaced with the new infected
and mutated version of loser. This is easy to imple-
ment and effective as well. This keeps in consistency
with the underlying intuition of microbial evolution
where evolution is brought without death of parents.
Investigation into Mutation Operators for Microbial Genetic Algorithm
301

3 EVALUATION
Performance of any optimization algorithm is mea-
sured through some characteristic features like its ro-
bustness and precision. A set of standard benchmark
problems are usually used to analyze these features
for any algorithm. These functions are well known in
literature, and their qualitative properties and global
extremes are also known. The notion of robustness
may refer to a set of characteristics itself. This tells
how good the algorithm can perform with increas-
ing number of dimensions and difficulty as well as
how timely it can converge to optimal solution. The
term convergence may be interpreted in two mean-
ings, either all the population becoming just uniform
at any suboptimal location or when the evaluation hits
the desired optimal results. Both these situations are
closely related but different from each other. Preci-
sion of the algorithm tells how close the results are to
desired optimal solution. A fast, robust and, precise
algorithm is an objective of optimization researcher.
A number of simulations are carried out to testify the
proposed suggestions. These simulations are run on
MATLAB 7.1 to give numerical results and graph-
ical representation of algorithm performances. We
have used a set of standard benchmark from literature
(Ortiz-Boyer et al., 2005) for our simulations catego-
rized under three subsets as follows.
• Unimodal multidimensional convex problems
• Multimodal two dimensional problems
• Multimodal multidimensional problems
Unimodal multidimensional convex problems include
some cases causing poor or slow convergenceto a sin-
gle global extreme. Multimodal problems may have
large number of local extremes making a multidimen-
sional problem harder to locate the global optima. In-
creasing number of dimensions further increases the
hardness of the problem but most of the real practical
problems desired to be modelled usually are multidi-
mensional. For evaluation, we run monte carlo sim-
ulations to find suitable controlling parameters like
probabilities and constants and then average the re-
sults from 100 simulations for each problem and vari-
ant of MGA.
3.1 Simulations Results
In this section, we have presented simulation plots
for MGA with different mutation operators. We have
used ranked roulette selection scheme in all simula-
tions and different mutation operators for each opti-
mization problem. The results plotted are mean val-
ues of 100 simulations for each evaluation. Proba-
bility of mutation and infection have been chosen af-
ter extensive experimental investigation. Table 1 pro-
vides chosen values for probabilities of infection Pi
and probability of mutation Pm for each problem and
also states the best solution achieved over 10000 it-
erations and n = 2, 30. MGA with adaptive mutation
is also compared with basic GA over 5000 iterations
using MATLAB GA toolbox with suggested values
of parameters from literature and ranked roulette se-
lection scheme as in MGA. Comparison with GA is
given in table 3 in appendix and shows that MGA
with adaptive mutation is highly comparable with ba-
sic GA and even outperforms GA in many problems
in terms of finding better minima specially in prob-
lems with less variables or dimensions.
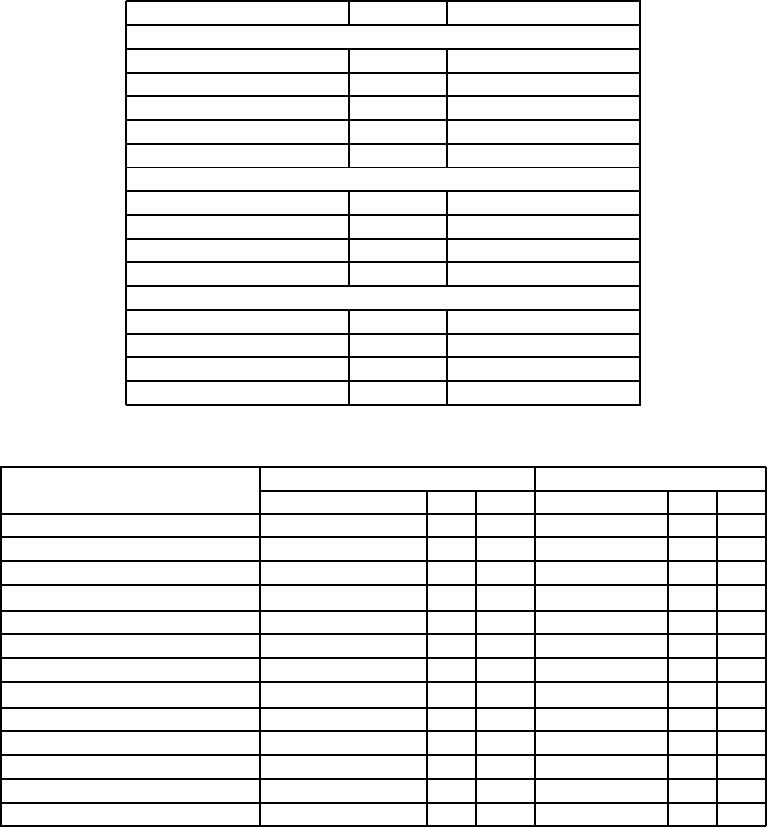
Table 1: Chosen Parameters and Best Achieved Solutions
in 5000 iterations.
Problem Best Minima Pi Pm
DeJong First 0.0002 0.7 0.7
Axis Parallel Ellipsoid 0.0025 0.7 0.7
Rotated Hyper Ellipsoid 0.0020 0.7 0.7
Sum of Different Powers 0.0092 x10
−4
0.7 0.7
Rosenbrok Valley 0.1081 0.7 0.7
Rastrigins 0.0329 0.7 0.7
Schwefel -1256.9 0.6 0.8
Griewangk 0.0566x10
−6
0.7 0.8
Ackley 0.0104 0.6 0.8
Branin 0.3978 0.5 0.75
Goldstein Price 3 0.6 0.4
Six Hump Camel Back -1.036 0.6 0.75
DeJong Fifth 0.998 0.6 0.75
3.1.1 Unimodal Multidimensional Problems
No. of Iterations
0 1000 2000 3000 4000 5000 6000 7000
Minima
×10
-3
0
0.5
1
1.5
2
2.5
3
Sum of Different Powers function
MGA
MGA-G
MGA-C
MGA-A
Figure 2: Sum of different powers function.
iterations
0 500 1000 1500 2000 2500 3000 3500 4000 4500 5000
optima
0
5
10
15
20
25
30
35
40
45
Axis Parallel Hyper Ellipsoid Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 3: Axis parallel hyper ellipsoid function.
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
302

iterations
0 1000 2000 3000 4000 5000 6000
optima
0
1
2
3
4
5
6
7
8
9
10
Dejong First Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 4: Dejong first function.
iterations
0 500 1000 1500 2000 2500 3000 3500 4000 4500 5000
optima
0
10
20
30
40
50
60
70
80
90
Rotated Hyper Ellipsoid Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 5: Rotated hyper ellipsoid function.
iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0
5
10
15
20
25
30
35
40
Rosenbrok Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 6: Rosenbrok valley function.
3.1.2 Multimodal Multidimensional Problems
No. of Iterations
0 1000 2000 3000 4000 5000 6000 7000
Minima
-14000
-12000
-10000
-8000
-6000
-4000
-2000
0
2000
4000
Schwefel function
MGA
MGA-G
MGA-C
MGA-A
Figure 7: Schwefel function.
iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0
2
4
6
8
10
12
Ackley Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 8: Ackley function.
iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0
5
10
15
20
25
30
35
Griewangk Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 9: Griewangk function.
iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0
20
40
60
80
100
120
Rastrigin Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 10: Rastrigin function.
3.1.3 Two Dimensional Multimodal Difficult
Problems
iterations
0 500 1000 1500 2000 2500 3000 3500 4000 4500 5000
optima
-1.032
-1.03
-1.028
-1.026
-1.024
-1.022
-1.02
Six Hump Camel Back Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 11: Six hump camel back function.
Investigation into Mutation Operators for Microbial Genetic Algorithm
303

iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0.39
0.4
0.41
0.42
0.43
0.44
0.45
0.46
0.47
0.48
0.49
Branin Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 12: Branin function.
iterations
0 1000 2000 3000 4000 5000 6000 7000 8000 9000 10000
optima
0
5
10
15
Dejong fifth Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 13: Dejong fifth function.
iterations
0 500 1000 1500 2000 2500
optima
0
10
20
30
40
50
60
70
80
Goldstein Price Function
MGA-uniform
MGA-Gaussian
MGA-Cauchy
MGA-Adaptive
Figure 14: Goldstein price function.
4 CONCLUSIONS
In this paper, we have analyzed performance of MGA
in minimizing unconstrained complex problems us-
ing commonly used mutation operators. We have also
suggested a simple adjustment to adapt the mutation
according to the diversity in population to be more
effective. Comparison of different variants of MGA
might be helpful for readers in choosing the suitable
variant according to their objective problem. MGA
with our adaptive mutation has shown promising be-
haviour in general and comparison with basic GA ver-
ifies its potential as a useful optimization technique.
REFERENCES
Al Jadaan, Omar, L. R. and Rao, C. (2008). Improved selec-
tion operator for genetic algorithm. Theoretical and
Applied Information Technology, (4.4).
Back, T. and Schwefel, H.-P. (1993). An overview of evo-
lutionary algorithms for parameter optimization. Evo-
lutionary computation, 1(1):1–23.
Baker, J. E. (1985). Adaptive selection methods for genetic
algorithms. In Proceedings of an International Con-
ference on Genetic Algorithms and their applications,
pages 101–111. Hillsdale, New Jersey.
Fogel, L. J., Owens, A. J., and Walsh, M. J. (1966). Artifi-
cial intelligence through simulated evolution.
Goldberg, D. E. and Deb, K. (1991). A comparative anal-
ysis of selection schemes used in genetic algorithms.
Foundations of genetic algorithms, 1:69–93.
Harvey, I. (2011). The microbial genetic algorithm. In Ad-
vances in artificial life. Darwin Meets von Neumann,
pages 126–133. Springer.
Hatwagner, F. and Horvath, A. (2012). Maintaining genetic
diversity in bacterial evolutionary algorithm. Annales
Univ. Sci. Budapest, Sec. Comp, 37:175–194.
John, H. (1992). Holland, adaptation in natural and artificial
systems: An introductory analysis with applications to
biology, control and artificial intelligence.
McCarthy, J. (2007). What is artificial intelligence. URL:
http://www-formal. stanford. edu/jmc/whatisai. html,
page 38.
Miller, B. L. and Goldberg, D. E. (1995). Genetic algo-
rithms, tournament selection, and the effects of noise.
Complex Systems, 9(3):193–212.
Ortiz-Boyer, D., Herv´as-Mart´ınez, C., and Garc´ıa-Pedrajas,
N. (2005). Cixl2: A crossover operator for evolution-
ary algorithms based on population features. J. Artif.
Intell. Res.(JAIR), 24:1–48.
Whitley, L. D. et al. (1989). The genitor algorithm and se-
lection pressure: Why rank-based allocation of repro-
ductive trials is best. In ICGA, pages 116–123.
Yao, X., Liu, Y., and Lin, G. (1999). Evolutionary program-
ming made faster. Evolutionary Computation, IEEE
Transactions on, 3(2):82–102.
ECTA 2015 - 7th International Conference on Evolutionary Computation Theory and Applications
304
APPENDIX
Table 2: Test Functions Used.
Name Min domain
Unimodal Multidimensional Problems
De Jong First 0 −5.12 ≤ x
i
≤ 5.12
Axis Parallel Ellipsoid 0 −5.12 ≤ x
i
≤ 5.12
Rotated Hyper Ellipsoid 0 −65.54 ≤ x
i
≤ 65.54
Sum of Different Powers 0 −1 ≤ x
i
≤ 1
Rosenbrocks Valley 0 −2.048 ≤ x
i
≤ 2.048
Multimodal Multidimensional Problems
Rastrigins 0 −5.12 ≤ x
i
≤ 5.12
Schwefel -418.98n −500 ≤ x
i
≤ 500
Grienwangk 0 −600 ≤ x
i
≤ 600
Ackley 0 −32.76 ≤ x
i
≤ 32.76
Difficult Two dimensional Multimodal Problems
Branin 0.397887 −5 ≤ x
i
≤ 15
Goldstein Price 3 −2 ≤ x
i
≤ 2
Six Hump Camel Back -1.036 −3 ≤ x
i
≤ 3
De Jong Fifth 0.998 −65.54 ≤ x
i
≤ 65.54
Table 3: Comparison of GA with MGA-Adaptive over 5000 iterations.
Problem MGA-Adaptive GA
Minima Achieved P
i
P
m
Minima P
c
P
m
DeJong First 0.0002 0.7 0.7 0.0001 0.8 0.2
Axis Parallel Hyper Ellipsoid 0.0025 0.7 0.7 0.0028 0.8 0.2
Rotated Hyper Ellipsoid 0.0020 0.7 0.7 0.0021 0.8 0.2
Sum of Different Powers 0.0092 x10
{−4}
0.7 0.7 0.0009 0.8 0.2
Rosenbrok Valley 0.1081 0.7 0.7 0.0541 0.8 0.2
Rastrigins 0.0329 0.7 0.7 0.004 0.8 0.1
Schwefel -1256.9 0.6 0.8 -1494.0 0.8 0.1
Griewangk 0.0566x10
{−6}
0.7 0.8 1.364x10
{−7}
0.8 0.1
Ackley 0.0104 0.6 0.8 0.0014 0.8 0.1
Branin 0.3960 0.5 0.75 0.3979 0.6 0.2
Goldstein Price 3.0000 0.6 0.4 3.002 0.6 0.2
Six Hump Camel Back -1.0360 0.6 0.75 -1.032 0.6 0.2
DeJong Fifth 0.9980 0.6 0.75 0.998 0.6 0.2
Investigation into Mutation Operators for Microbial Genetic Algorithm
305
