
Gene Selection using a Hybrid Memetic and Nearest Shrunken
Centroid Algorithm
Vinh Quoc Dang and Chiou-Peng Lam
School of Computer & Security Science, Edith Cowan University, Perth, Australia
Keywords: Memetic Algorithm, Nearest Shrunken Centroid, Microarray, Feature Selection, Classification.
Abstract: High-throughput technologies such as microarrays and mass spectrometry produced high dimensional
biological datasets both in abundance and with increasing complexity. Prediction Analysis for Microarrays
(PAM) is a well-known implementation of the Nearest Shrunken Centroid (NSC) method which has been
widely used for classification of biological data. In this paper, a hybrid approach incorporating the Nearest
Shrunken Centroid (NSC) and Memetic Algorithm (MA) is proposed to automatically search for an optimal
range of shrinkage threshold values for the NSC to improve feature selection and classification accuracy.
Evaluation of the approach involved nine biological datasets and results showed improved feature selection
stability over existing evolutionary approaches as well as improved classification accuracy.
1 INTRODUCTION
Recent reviews (Hilario and Kalousis, 2008) have
described numerous feature selection techniques for
identifying informative biomarkers from biological
datasets. The two main objectives of feature
selection is achieving high classification accuracy
and high reproducibility of a pertinent list of
biomarkers (i.e. feature selection stability). Stability
is a term used to describe the sensitivity of a feature
selection algorithm to small variations in the training
data and in the settings of the algorithmic
parameters, resulting in different feature sets being
produced by the algorithm.
Many studies (Kim et al., 2010; Yu and Liu,
2004), have used the Nearest Shrunken Centroid
(NSC) algorithm (Tibshirani et al., 2002) for feature
selection (FS) and classification in high dimensional
biomedical data. This algorithm, with its most well-
known software implementation being known as
Prediction Analysis for Microarrays (PAM), requires
a shrinkage threshold value as input for performing
FS and classification. The choice of this threshold
value, as stated in the PAM User guide, is
determined “after a judicious examination of
training errors and the cross-validation results”.
Hence, the selection of the optimal shrinkage
threshold value is typically a manual process based
on “trial and error” by setting the shrinkage
threshold value to vary equally using a predefined
step size across a predefined range (Lusa, 2012).
However, shrinkage threshold values selected in this
way may not give optimal solutions (Dang et al.,
2013) and is also a very time consuming process.
A hybrid approach (NSC-GA) (Dang et al.,
2013), incorporating GA and NSC to automatically
find the optimal shrinkage threshold value.
Computation time associated with GA processing
can be intensive (Elbeltagi et al., 2005) . One of the
approaches to improve GAs both in terms of
computation time and quality of optimal solutions is
the use of a memetic algorithm (MA) (Elbeltagi et
al., 2005).
In this paper, an approach of incorporating the
NSC algorithm into a MA, namely NSC-MA, for
automatically searching for an optimal range of
shrinkage threshold values is proposed. The aim
here is to explore how to improve the NSC-GA
approach (Dang et al., 2013) for achieving
robustness of selected feature subsets and stability in
signatures of biomarkers. Unlike NSC-GA, the
proposed approach consistently reproduces the same
candidate feature subset from repeated runs
involving a dataset.
The rest of the paper is organized as follows:
Section 2 reviews some related work, Section 3
describes details of the proposed approach, datasets,
results and discussion are presented in Section 4, and
conclusion is drawn in Section 5.
190
Dang, V. and Lam, C-P.
Gene Selection using a Hybrid Memetic and Nearest Shrunken Centroid Algorithm.
DOI: 10.5220/0005665201900197
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 190-197
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 RELATED WORK
Chin et al., (2015) completed a comprehensive
review of feature selection methods for gene
selection, categorising these into three classes,
namely supervised, unsupevised and semi-
supervised. Each of these 3 classes are further
refined into sub-categories on the basis of evaluation
criterion into filter, wrapper or embedded methods.
Statistical metrics are used in filter methods to rank
each feature individually or subsets of features for
its ability to discriminate between classes. In
wrapper-based methods, classification models are
used to determine the relevance of sets of features
and embedded methods are similar to wrapper
methods except for a much tighter coupling between
feature selection and classifier. Feature selection is
NP-hard and can be approximated via a heuristic
search for an "optimal" feature subset. In conclusion,
Chin et al., (2015) discussed a number of areas
needing future research, amongst these, is the need
to develop methods for robustness of selected
feature subsets (i.e. stability of signature).
Dang et al., (2013) developed a wrapper
approach (NSC-GA) involving genetic algorithm
and NSC and evaluated the approach on microarray
data. Similar to PAM (Tibshirani et al., 2002), the
selection of subsets of features utilize a penalized t-
statistic but the approach automatically determines
the required soft-threshold for identifying a gene
set for classification. Experimental results show that
the optimal threshold value obtained using NSC-GA
resulted in a smaller number of features and higher
classification accuracies on test datasets in
comparison to previous studies such as Klassen and
Kim (2009).
Soufan et al., (2015) developed a web-based,
wrapper feature selection tool using a parallel GA as
its search strategy that allows concurrent evaluations
of large number of candidate subsets. The tool is
flexible for its range of filtering methods as well as
its functionality of allowing for adjustments of
weights and parameters in the fitness function.
Zhu et al., (2007b) incorporated a memetic
algorithm (MA) in their approaches, namely
WFFSA and MBEGA (Zhu et al., 2007a) for finding
relevant features in microarray data. Both these
approaches were based on the traditional GA and a
local search (LS) algorithm that incorporated filter
ranking method for WFFSA and Markov Blanket for
MBEGA respectively. Binary representation (1, 0)
was the encoding for individuals and the SVM
classifier was employed to evaluate the fitness of
individuals. Empirical evaluations of these two
approaches on microarray datasets indicate that they
outperformed many existing methods in terms of
classification accuracy, number of selected genes
and search efficiency.
3 NSC-MA PROPOSED
APPROACH
MA is a hybrid of EAs which involves an
evolutionary algorithm (EA) and a local search (LS)
to improve the fitness of chromosome (Krasnogor
and Smith, 2005; Wu, 2001). As shown in Figure 1,
the 2 major steps in NSC-MA are:
Step 1: This step involved the automatic
calculation of Th
max
. This procedure is performed
once only at the beginning of the proposed approach,
NSC-MA, to obtain Th
max.
Step 2: MA (Moscato, 1989) is employed in this
step as an optimization method to search for optimal
sets of shrinkage thresholds for NSC algorithm that
lead to the selection of optimal sets of features. NSC
algorithm is employed as a fitness evaluator to
evaluate the fitness of each chromosome in terms of
the number of selected features and its
corresponding training classification accuracy.
Figure 1: Framework of the proposed approach, NSC-MA,
using MA with adding and subtracting Improvement First
Strategy LS.
Gene Selection using a Hybrid Memetic and Nearest Shrunken Centroid Algorithm
191

Gene value in each chromosome in the population is
initialized to a real value within the range of [0,
Th
max
] using a random number generator. The
random number generator uses a different seed for
each initialization of a new population. Details
associated with determination of Th
max
can be found
in Dang et al., (2013).
3.1 Fitness Evaluation
The NSC algorithm (Tibshirani et al., 2002) is the
fitness evaluator for obtaining the overall fitness,
Fitness
Ind
, for each individual chromosome. As
defined in Equation (1), Fitness
Ind
is calculated by
averaging the fitness values associated with all the
threshold values for a chromosome.
Fitness
In
d
=
∑
f
th
/ M
(1)
where M is a number of genes or threshold values in
a chromosome.
The function f
th
in Equation (2) consists of two
other functions, f
1
and f
2
:
f
th
= f
1
+ f
2
(2)
f
1
= (N
total
- N
att
) / N
total
(3)
f
2
=
TP+TN
TP+FP+TN+FN
(4)
where TP is the true positives, TN is the true
negative, FP is the false positive, FN is the false
negative. N
total
equals to the total number of
attributes (features) in the dataset, N
att
is the number
of attributes selected by NSC. f
1
is designed for
evaluating the fitness of a threshold that leads to a
minimum number of attributes, whilst f
2
is
associated with the maximum classification
accuracy.
3.2 Generating New Population
The procedure for generating a new population using
MA incorporated adding and subtracting LS with
Improvement First Strategy is as follows:
Input:
Chromosome population (p)
Fitness population (F
p
)
Crossover probability (P
c
)
Mutation probability (P
m
)
Elite chromosome (Elite)
Chromosome length (len
C
)
Output:
New population (N
p
)
Steps:
1. Set Size=size of population, p
2. Set new population (N
p
)={∅}
3. Store Elite into N
p
4. For counter from 1 to ½ Size
a. Select 2 parent chromosomes
using binary tournament selection
i. Select 2 chromosomes randomly
from p
Select the best fit chromosome as
1
st
parent (parent
1
)
ii. Select 2 chromosomes randomly from
p
Select the best fit chromosome as
2
nd
parent (parent
2
)
b. Create 2 offspring chromosomes using
parent
1
and parent
2
i. Generate a random number (R
n
) in
the range [0, 1] using RNG
ii. If R
n
≤ P
c
Perform one point crossover on 2
parents to produce offspring
1
and
offspring
2
Perform adding and subtracting LS
with Improvement First Strategy
on offspring
1
and offspring
2
to
produce 2 new offspring
(offspring
1
ls
cross
and
offspring
2
ls
cross
)
iii. If R
n
≤ P
m
For counter from 1 to len
C
Generate a random number
(R
n
) in the range [0, 1]
using RNG
If R
n
≤ P
m
Perform uniform mutation on
each bit of offspring
1
to
generate offspring
1mut
Perform uniform mutation on
each bit of offspring
2
to
generate offspring
2mut
Perform adding and
subtracting LS with
Improvement First Strategy
on offspring
1mut
and
offspring
2mut
to produce 2
new offspring
(offspring
1
ls
mut
and
offspring
2
ls
mut
)
iv. Evaluate fitness of
offspring
1
ls
cross,
offspring
2
ls
cross
,
offspring
1
ls
mut
and offspring
2
ls
mut
chromosomes
c. Store the best 2 chromosomes into N
p
.
This step involved the “adding and subtracting LS
with Improvement First Strategy” step, which is
applied to offspring chromosomes after crossover
and mutation in order to further improve its quality.
A single elitist strategy is employed where the
best candidate solution (elite) from the previous
generation is placed into the new population. To
produce new offspring, Binary Tournament selection
is used to select the individuals as parents to go
through crossover, mutation and LS strategy. Two
best offspring chromosomes from each of these
iterations are then placed into the new population.
The procedure for the “adding and subtracting
LS with Improvement First Strategy” step is as
follows:
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
192
Input:
Chromosome (chrom)
Chromosome length (len)
Output:
An improved local search chromosome
(chrom
ls
)
Steps:
1. Generate a real random number (R
n
)
in the range [0,1] using RNG
2. Evaluate fitness of chrom
3. set fitness of chrom
ls
=0
4. set counter=1
5. While (counter<=len) and (fitness
chrom
ls
<=fitness chrom)
a. Add R
n
to chrom[counter] to
create a new chromosome (chrom
ls
)
b. Evaluate the fitness of chrom
ls
c. If fitness of chrom
ls
> chrom
Retain chrom
ls
as an improved
local search chromosome
d. Else
subtract R
n
to chrom[counter]
create a new chromosome
(chrom
ls
)
evaluate the fitness of chrom
ls
If fitness of chrom
ls
> chrom
retain chrom
ls
as an improved
local search chromosome
Else
discard chrom
ls
update counter=counter+1
3.3 Parameter Settings
Table 1: Parameter settings used in the proposed approach
NSC-MA.
Parameters Values/Algorithm
Population size 30
Chromosome length 10
Crossover rate 0.6
Mutation rate 0.0333
Maximum generation 1000
Selection Binary Tournament
Crossover Single point
Mutation Uniform
Elitist Single
Local search
Adding and subtracting with First
Improvement Strategy
The parameter settings for running NSC-MA are
shown in Table 1. The parameters that are tuned
include population size, crossover probability rate,
and mutation probability rate, with these values in
the table, being taken from an empirical experiment
(Dang, 2014). Uniform mutation (Eiben and Smith,
2007) modifies a chromosome by replacing its gene
value with a mutated number, N
mut
, which is
calculated using equation (5).
N
mu
t
= L
b
+ (R
n
* (U
b
- L
b
)) (5)
where L
b
is lower bound of chromosome, R
n
is a
random number generated by RNG, U
b
is upper
bound of chromosome.
4 RESULTS AND DISCUSSION
Table 2 showed a summary of the nine datasets that
have been used widely by many recent
investigations as demonstrated in Chin et al (2015).
These include: AD Disease (Ray et al., 2007), Colon
(Alon et al., 1999), Leukemia (Golub et al., 1999),
Ovarian (Petricoin et al., 2002), Lymphoma
(Alizadeh et al., 2000), Lung (Gordon et al., 2002),
Prostate (Singh et al., 2002), Central Nervous
System (CNS
) (Pomeroy et al., 2002) and Breast-
A (van't Veer et al., 2002) that we used to evaluate
NSC-MA. Each dataset is partitioned into a training
dataset and an unseen test set using either the same
configuration as proposed by their original authors
(as cited for each dataset mentioned above), or those
of other authors who have used the same datasets in
their studies.
Table 2: Summary of nine public datasets used for the
NSC-MA approach.
Dataset Type of data
No of
attr.
No of
classes
No of
Samples
AD
Protein
Immunoa-ssay
120 2 259
Colon
Cancer
microarray
2000 2 62
Leukemia 7129 2 72
Lung 12533 2 181
Lymphoma 4026 2 47
Prostate 12600 2 136
CNS 7129 2 60
Breast-A 1213 3 98
Ovarian
Proteomic
spectra
15154 2 253
For each of the nine datasets, 15 independent
runs of NSC-MA were executed using the respective
training dataset and parameter values shown in
Table 1. Each independent run involved an initial
population produced using the Random Number
Generator with a random seed. For each run, 10 fold
cross validation (CV) strategy was employed to
evaluate the selected feature sets. The optimal set of
features was then used to construct the NSC
classifier to classify the unseen test data associated
with the dataset. The average classification accuracy
was calculated from these runs.
A simple multi-start local search algorithm
(MSLS) based on a local search method (Lourenço
et al., 2001) was implemented for comparison of
performance with NSC-MA. 15 independent runs of
Gene Selection using a Hybrid Memetic and Nearest Shrunken Centroid Algorithm
193

MSLS were also executed using the respective
training dataset.
The results are examined from 2 perspectives:
diagnostic relevance in terms of features used in the
construction of accurate diagnostic classifiers for
prediction and by examination of the literature for
the established implication of the selected set of
features to specific diseases (Table 4). Table 3
showed comparisons of results from NSC-MA with
MSLS and other studies using equivalent protocol,
that is training a classifier using a training set and
evaluation of performance involved an unseen test
set. NSC-MA consistently selected only one set
features over 15 independent runs. This shows that
the stability of NSC-MA is improved over NSC-GA.
For example for Colon cancer dataset, NSC-GA
selected 2 sets of 6 and 28 features, whilst NSC-MA
selected only one set of 28 features, for Lung cancer
dataset, NSC-GA selected 4 sets of 8, 9, 10 and 11
features whilst NSC-MA selected only one set of 8
features with the same classification accuracy of
100%. With the AD, CNS and Breast-A datasets,
both NSC-MA and MSLS returned the same results
but with the remaining 6 datasets, there is a lot more
variability in terms of the number of selected subsets
as well as the number of features in the respective
feature subsets from employing MSLS, thus
demonstrating that NSC-MA has better feature
selection stability over MSLS.
NSC-MA achieved very similar classification
results to NSC-GA. In comparison to other existing
techniques, NSC-MA achieved better classification
results in most cases using a smaller feature sets.
The set of 11 features associated with the AD dataset
is a subset of the 18 identified by Ray et al., (2007).
For the Colon dataset, it is not possible to check the
set of 28 genes found by the proposed approach
against the set of 16 genes in Klassen and Kim
(2009) as these were not listed in their study.
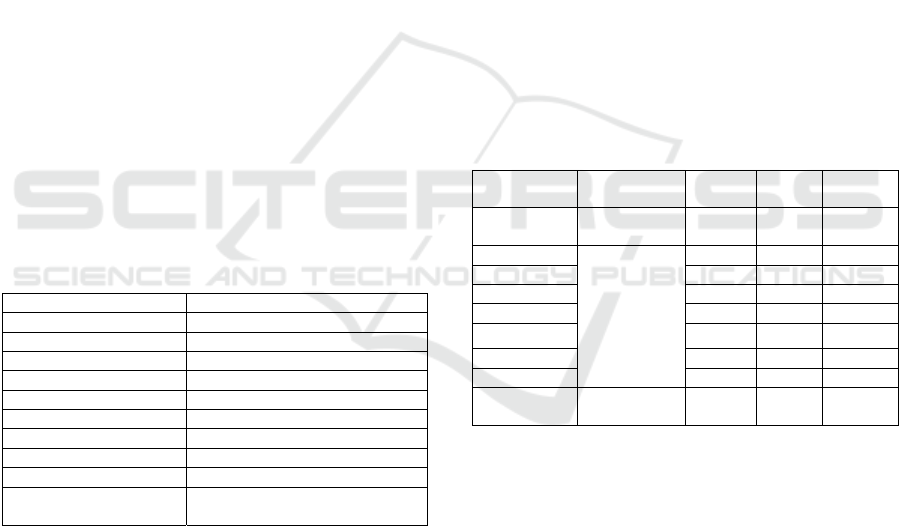
Table 3: Summary of results obtained from the NSC-MA approach in comparison with existing approaches. Each cell
indicates the average unseen test classification % and the number of selected genes in () associated with 15 independent
runs. In cells with multiple entries, this is associated with some of the 15 runs returning different subsets of features.
Approach AD Colon Leukemia Ovarian Lymphoma Lung Prostate CNS Breast-A
Proposed
approach
NSC-MA
89.34
(11)
100
(28)
97.05
(9)
96.06
(7)
100(128) 100(8) 90.2(6) 65.51(3) 89.58(2)
NSC-GA(Dang
et al., 2013)
89.49
(11)
93.75
(6)
100
(28)
97.05
(9)
96.06
(7)
95.45(7)
95.45(12)
100(128)
100(129)
100(132)
100(8)
100(9)
100(10)
100(11)
90.2(6) 65.51(3) 89.58(2)
NSC (Ray et
al., 2007)
89
(18)
NSC (Klassen
and Kim, 2009)
75(16)
94.12
(21)
86.6(25) 93.7(5) 90.91(6)
ALP-NSC,
AHP-NSC
(Wang and Zhu,
2007)
94.12
(16)
Weighted NSC
(Tai and Pan,
2007)
99.55(6)
60.51
(10)
FAIR (Gordon
et al., 2002)
97.05
(11)
95.3(31) 73.52(2)
GCLUS &
SERA
(Baggiolini et
al., 1989)
97.63
(47)
Multi-Start
Local Search
(MSLS)
88.62
(11)
93.75 (1)
93.75(6)
100(28)
93.75(29)
87.5(34)
87.5(35)
91.17(1)
91.17(2)
91.17(3)
96.06(7)
96.06(36)
96.06(37)
96.06(38)
95.45(7)
100
(128, 130, 132,
135, 137, 139,
145, 140, 151)
100(8)
100(9)
100(10)
100(11)
88.23(3)
88.23(4)
90.2(5)
90.2(6)
65.51(3)
89.58
(2)
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
194

Table 4: The sets of features selected by NSC-MA for nine datasets AD, Colon, Leukemia, Ovarian, Lymphoma, Lung,
Prostate CNS and Breast-A.
Dataset
No of
Attr
Acc No
AD 11
PDGF-BB_1 RANTES_1 IL-1a_1 TNF-a_1 EGF_1 M-CSF_1 ICAM-1_1 IL-11_1 IL-
3_1 GCSF_1 ANG-2_1
Colon 28
T95018, X55715, M63391, H40560, T92451, T57619, R78934, T58861, M26697,
M76378, R87126, H43887, H64489, M22382, T71025, Z24727, Z50753, X12671,
T47377, L05144, H55758, M64110, M76378, T60155, M76378, J02854, X86693,
T60778
Leukemia 9 M27891, M84526, M96326, U46751, U50136, X17042, X95735, M28310, Y00787
Ovarian 7
MZ244.36855, MZ244.66041, MZ244.95245, Z245.24466, MZ245.8296,
MZ245.53704 and MZ246.12233
Lymphoma 7
GENE3327X, GENE3329X, GENE3330X, GENE3332X, GENE3361X,
GENE3258X, GENE3256X
Lung 8 32551_at, 33328_at, 34320_at, 36533_at, 37157_at, 37716_at, 37954_at, 40936_at
Prostate 6 31444_s_at, 41468_at, 37639_at, 38406_f_at, 769_s_at and 556_s_at
CNS 3 L17131_rna1_at, Yo7604_at, U33448_s_at
Breast-A 2 LY6D, ESR1
In the case of the Leukemia cancer dataset, NSC-
MA obtained a smaller set of 9 genes and
classification accuracy of 97.05% when compared
to 96% using 10 genes in (Huang, 2009) with 2
genes (M27891, X95735) in common. Eight genes
(M27891, M84526, M96326, U46751, U50136,
X95735, M28130, Y00787) are a subset of the set of
48 features selected using GA and ANNs in (Tong et
al., 2009).
This nine gene is also a subset of the set of 50
highly expressed genes identified by (Masys et al.,
2001) for predicting disease from non-disease. For
the Ovarian cancer dataset, NSC-MA identified a set
of 7 features, MZ244.36855, MZ244.66041,
MZ244.95245, Z245.24466, MZ245.8296,
MZ245.53704 and MZ246.12233 which is a subset
of the 47 peptides reported in Foss (Foss, 2011),
with similar classification accuracy of 96.06% on
the unseen test set. Six peptides in this set are among
the top 10 peptides identified in Yap et al., (2007).
In terms of the Lung cancer dataset, using a set of 6
features, Tai and Pan (2007) achieved 99.55%
whereas NSC-MA used 8 features and obtained
100% classification accuracy on the unseen test set.
However, the identified features have not been listed
in Tai and Pan’s paper. For the Breast cancer
dataset, NSC-MA identified a set of 2 features,
LY6D (Lymphocyte antigen 6 complex, locus D)
and ESR1 (Estrogen receptor 1). LY6D is strongly
expressed in cervical cancer, head and neck cancer,
lung cancer, skin cancer and urothelial cancer, and
also a marker of the earliest stage of B-cell
specification (GeneCards; The Human Protein
Atlas). ESR1 is cancer and disease related genes,
and also involved in pathological processes in
endometrial and breast cancer (The Human Protein
Atlas).
To obtain an overall estimate of the
computational effort of using NSC-GA and NSC-
MA to analyse the nine datasets, we collected the
total time taken for each of their 15 independent
runs. The average time taken by NSC-GA is 1290.39
minutes and 1219.56 minutes for NSC-MA.
5 CONCLUSIONS
The shrinkage threshold value must be provided as
an input to the NSC algorithm and an appropriate
choice is extremely important in terms of feature
selection and classification accuracy. Researchers
have used approaches of trial and error to select a
threshold value that produced minimum
classification errors and some emerging work has
investigated approaches to automatically produce
this value. A novel approach incorporating NSC and
MA algorithm is proposed in this study in order to
overcome limitations of the previous approaches
such as empirical methods with NSC and NSC-GA.
Evaluation of the approach involved nine biological
datasets and results showed improved feature
selection stability over existing evolutionary
approaches as well as improved classification
accuracy.
REFERENCES
Alizadeh, A., Eisen, B., Davis, E., Ma, C., Lossos, I. S.,
Rosenwald, A., Yu, X. (2000). Distinct types of
Gene Selection using a Hybrid Memetic and Nearest Shrunken Centroid Algorithm
195

diffuse large B-cell lymphoma identified by gene
expression profiling. Nature, 403(6769), 503-511.
Alon, Uri, Barkai, Naama, Notterman, Daniel A, Gish,
Kurt, Ybarra, Suzanne, Mack, Daniel, & Levine,
Arnold J. (1999). Broad patterns of gene expression
revealed by clustering analysis of tumor and normal
colon tissues probed by oligonucleotide arrays.
Proceedings of the National Academy of Sciences,
96(12), 6745-6750.
Baggiolini, Marco, Walz, A, & Kunkel, SL. (1989).
Neutrophil-activating peptide-1/interleukin 8, a novel
cytokine that activates neutrophils. Journal of Clinical
Investigation, 84(4), 1045.
Chin, A., Mirzal, A., Haron, H., & Hamed, H. (2015).
Supervised, Unsupervised and Semi-supervised
Feature selection: A Review on Gene Selection.
Dang, V. (2014). Evolutionary approaches for feature
selection in biological data. (PhD), Edith Cowan
University, Australia.
Dang, V., Lam, C., & Lee, C. (2013). NSC-GA: Search for
optimal shrinkage thresholds for nearest shrunken
centroid. Paper presented at the Proceedings IEEE
sympodium series on computatinal intelligence,
Singapore.
Eiben, A. E., & Smith, J. E. (2007). Introduction to
evolutionary computing. Berlin Heidelberg: Springer.
Elbeltagi, Emad, Hegazy, Tarek, & Grierson, Donald.
(2005). Comparison among five evolutionary-based
optimization algorithms. Advanced Engineering
Informatics, 19(1), 43-53.
Foss, Andrew. (2011). High-dimensional Data Mining:
Subspace Clustering, Outlier Detection and
Applications to Classification: VDM Publishing.
GeneCards.). LY6D Gene. Retrieved 10 December,
2015, from http://www.genecards.org/cgi-
bin/carddisp.pl?gene=LY6D.
Golub, Todd R, Slonim, Donna K, Tamayo, Pablo, Huard,
Christine, Gaasenbeek, Michelle, Mesirov, Jill P,
Caligiuri, Mark A. (1999). Molecular classification of
cancer: class discovery and class prediction by gene
expression monitoring. Science, 286(5439), 531-537.
Gordon, Gavin J, Jensen, Roderick V, Hsiao, Li-Li,
Gullans, Steven R, Blumenstock, Joshua E,
Ramaswamy, Sridhar, . . . Bueno, Raphael. (2002).
Translation of microarray data into clinically relevant
cancer diagnostic tests using gene expression ratios in
lung cancer and mesothelioma. Cancer Research,
62(17), 4963-4967.
Hilario, M., & Kalousis, A. (2008). Approaches to
dimensionality reduction in proteomic biomarker
studies. Briefings in Bioinformatics, 9(2), 102-118.
Huang, Liang-Tsung. (2009). An integrated method for
cancer classification and rule extraction from
microarray data. J Biomed Sci, 16(1), 25.
Kim, Gilhan, Kim, Yeonjoo, Lim, Heuiseok, & Kim,
Hyeoncheol. (2010). An MLP-based feature subset
selection for HIV-1 protease cleavage site analysis.
Artificial Intelligence in Medicine, 48(2-3), 83-89.
Klassen, M., & Kim, N. (2009). Nearest shrunken centroid
as feature selection for microarray data.
Paper
presented at the ICATA (Computers and Their
Applications).
Krasnogor, Natalio, & Smith, Jim. (2005). A tutorial for
competent memetic algorithms: model, taxonomy, and
design issues. Evolutionary Computation, IEEE
Transactions on, 9(5), 474-488.
Lourenço, Helena R, Martin, Olivier C, Stützle, Thomas,
Glover, Ed F, & Kochenberger, G. (2001). Iterated
Local Search. arXiv preprint math.OC/0102188.
Lusa, Lara. (2012). Impact of class-imbalance on multi-
class high-dimensional class prediction. Metodoloski
zvezki, 9(1), 25.
Masys, Daniel R, Welsh, John B, Fink, J Lynn, Gribskov,
Michael, Klacansky, Igor, & Corbeil, Jacques. (2001).
Use of keyword hierarchies to interpret gene
expression patterns. Bioinformatics, 17(4), 319-326.
Moscato, Pablo. (1989). On evolution, search,
optimization, genetic algorithms and martial arts:
Towards memetic algorithms. Caltech concurrent
computation program, C3P Report, 826, 1989.
Petricoin, EF, Ardekani, AM, Hitt, BA, Levine, PJ,
Fusaro, VA, Steinberg, SM, Liotta, LA. (2002). Use of
proteomic patterns in serum to identify ovarian cancer.
The Lancet, 359(9306), 572 - 577.
Pomeroy, SL, Tamayo, P, Gaasenbeek, M, Sturla, LM,
Angelo, M, McLaughlin, ME, Golub, TR. (2002).
Prediction of central nervous system embryonal
tumour outcome based on gene expression. Nature,
415, 436 - 442.
Ray, Sandip, Britschgi, Markus, Herbert, Charles, Takeda-
Uchimura, Yoshiko, Boxer, Adam, Blennow, Kaj,
Karydas, Anna. (2007). Classification and prediction
of clinical Alzheimer's diagnosis based on plasma
signaling proteins. Nature medicine, 13(11), 1359-
1362.
Singh, Dinesh, Febbo, Phillip G, Ross, Kenneth, Jackson,
Donald G, Manola, Judith, Ladd, Christine, Richie,
Jerome P. (2002). Gene expression correlates of
clinical prostate cancer behavior. Cancer Cell, 1(2),
203-209.
Soufan, O, Kleftogiannis, D, Kalnis, P, & Bajic, VB.
(2015). DWFS: A Wrapper Feature Selection Tool
Based on a Parallel Genetic Algorithm. PLoS ONE,
10(2), e0117988.
Tai, F., & Pan, W. (2007). Incorporating prior knowledge
of gene functional groups into regularized discriminant
analysis of microarray data. Bioinformatics, 23(23),
3170-3177.
The Human Protein Atlas.). ESR1. Retrieved 10
December, 2015, from http://www.proteinatlas.org/
ENSG00000091831-ESR1/gene.
Tibshirani, R., Hastie, T., Narasimhan, B., & Chu, G.
(2002). Diagnosis of multiple cancer types by
shrunken centroids of gene expression. Proc Natl
Acad Sci USA, 99(10), 6567 - 6572.
Tong, Dong L, Phalp, Keith T, Schierz, Amanda C, &
Mintram, Robert. (2009). Innovative hybridisation of
genetic algorithms and neural networks in detecting
marker genes for leukaemia cancer. Paper presented at
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
196

the 4th IAPR International Conference in Pattern
Recognition for Bioinformatics, Sheffield, UK.
van't Veer, Laura J, Dai, Hongyue, Van De Vijver, Marc J,
He, Yudong D, Hart, Augustinus AM, Mao, Mao, . . .
Witteveen, Anke T. (2002). Gene expression profiling
predicts clinical outcome of breast cancer. Nature,
415(6871), 530-536.
Wang, S., & Zhu, J. (2007). Improved centroids estimation
for the nearest shrunken centroid classifier.
Bioinformatics, 23(8), 972-979.
Wu, Fengjie. (2001). A Framework for Memetic
Algorithms. (Master of Science in Computer Science),
University of Auckland, Auckland.
Yap, E., Tan, H., & Pang, H. (2007). Learning causal
models for noisy biological data mining: An
application to ovarian cancer detection. Paper
presented at the AAAI.
Yu, L., & Liu, H. (2004). Redundancy based feature
selection for microarray data. Paper presented at the
Proceedings of the tenth ACM SIGKDD international
conference on Knowledge discovery and data mining.
Zhu, Z., Ong, Y., & Dash, M. (2007a). Markov blanket-
embedded genetic algorithm for gene selection.
Pattern Recognition, 40(11), 3236-3248.
Zhu, Z., Ong, Y., & Dash, M. (2007b). Wrapper–filter
feature selection algorithm using a memetic
framework. Systems, Man, and Cybernetics, Part B:
Cybernetics, IEEE Transactions on, 37(1), 70-76.
Gene Selection using a Hybrid Memetic and Nearest Shrunken Centroid Algorithm
197
