
BioMed Xplorer
Exploring (Bio)Medical Knowledge using Linked Data
Mohammad Shafahi, Hayo Bart and Hamideh Afsarmanesh
Informatics Institute, Faculty of Science, University of Amsterdam, Science Park 904, Amsterdam, The Netherlands
Keywords:
BioMed Xplorer, Disease Related Information, Semantic Web, Knowledge Base Ontology, Visualization,
Provenance Data, Medical Knowledge, External Data Source, RDF, Graph, Knowledge Exploration.
Abstract:
Developing an effective model for predicting risks of a disease requires exploration of a vast body of
(bio)medical knowledge. Furthermore, the continuous growth of this body of knowledge poses extra chal-
lenges. Numerous research has attempted to address these issues through developing a variety of approaches
and support tools. Most of these tools however, do not sufficiently address the needed dynamism, lack in-
tuitiveness in their use, and present a rather scarce amount of information usually obtained from a single
source. This research aims to address the aforementioned gaps through the development of a dynamic model
for (bio)medical knowledge, represented as a network of interrelated (bio)medical concepts, and integrating
disperse sources. To this end, this paper introduces BioMed Xplorer, presenting a model and a tool that enables
researchers to explore biomedical knowledge, organized in an information graph, through a user friendly and
intuitive interface. Furthermore, BioMed Xplorer provides concept related information from a multitude of
sources, while also preserving and presenting their provenance data. For this purpose a RDF knowledge base
has been created based on a core ontology which we have introduced. Results are further experimented with
and validated by some domain experts and are contrasted against the state of the art.
1 INTRODUCTION AND
RESEARCH APPROACH
The (bio)medical field is vast and dynamic, with
knowledge developing rapidly as a result of contin-
uously ongoing research. Within this field, extensive
research is conducted into identifying risk factors of
diseases as well as assessing their effect on the pres-
ence and associated severity of a disease. The avail-
able knowledge from this research on risk factors en-
ables researchers to develop models for risk predic-
tion, which might be used by practitioners to assess
someone’s risk on developing a particular disease.
Conventional methods for developing such models for
risk prediction would involve identifying the risk fac-
tors and their effects from the ever-evolving body of
(bio)medical knowledge. Achieving this aim would
thus involve checking vast amount of scientific publi-
cations for relevant statements regarding factors that
might affect a disease. This, however, is a cumber-
some and costly activity, especially when considering
the fact that the U.S. National Library of Medicine’s
(NLM) bibliographic database MEDLINE, as of to-
day, contains over 22 million citations, over 750,000
of which were added in 2014 (U.S. National Library
of Medicine, 2015b), and that these numbers have
grown exponentially (Hunter and Cohen, 2006). As
a result of the sheer size and continuous growth of
the body of (bio)medical knowledge, exploration of
this body of knowledge, as well as finding the relevant
knowledge for inclusion in models for risk prediction,
becomes increasingly challenging for researchers, po-
tentially causing an information overload (Hunter and
Cohen, 2006; Lu, 2011).
Numerous researchers have reckoned this prob-
lem and have attempted to address it from differ-
ent perspectives (Lu, 2011; Cohen and Hersh, 2005),
for example through the development of comprehen-
sive visualizations that represent knowledge extracted
from (bio)medical publications (Plake et al., 2006;
Rebholz-Schuhmann et al., 2007; Tao et al., 2005;
Kilicoglu et al., 2008; Bodenreider, 2000).
Even though the visual nature of these knowledge
representation and visualization tools provides them
with great expressive power, four common shortcom-
ings can be identified among them, being: i) their re-
stricted scope, focusing just on a particular sub-do-
main of the (bio)medical field, ii) their lack of in-
Shafahi, M., Bart, H. and Afsarmanesh, H.
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data.
DOI: 10.5220/0005700300510062
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 51-62
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
51

tuitiveness and rather sharp learning curve, iii) the
scarce amount of information represented, solely lim-
ited to names and identifiers, lacking descriptions
or definitions, while these are available externally,
and iv) the fact that they are either no longer ac-
tive (AliBaba, PGviewer), or do not work properly
(EBIMed). From these shortcomings it thus becomes
clear that there is a need for a meaningful represen-
tation of the available (bio)medical knowledge that:
a) is intuitive, and b) represents information from
multiple sources. As such the following research
question can be conceived:
Can we develop a model of the (bio)medical
knowledge that is available from large, disperse, het-
erogeneous, and dynamic sources across the web?
In order to address this research question a five-
phase research approach has been designed, consist-
ing of the following phases: 1) State of the Art As-
sessment, 2) Data Source Characterization and Se-
lection, 3) Data Preprocessing and Ontology Design,
4) Data Interlinking and Fusion with external sources,
and 5) Model Visualization.
Completion of these five phases delivers a system
with an architecture that is shown in Figure 1. As
one might notice, the architecture consists of four core
modules, each of which corresponds to one of the ma-
jor design and development stages. The components
of these modules will be gradually defined in the cor-
responding sections, as such fully describing the sys-
tem architecture.
The remainder of this paper is structured accord-
ing to the five phases that were outlined above, with
each section elaborately discussing a particular phase
of the research. In section 2, the characterization and
selection of data sources for inclusion in the model
is described. This is followed by a discussion of the
data preprocessing and ontology design in section 3,
whereas section 4 covers the fusion and interlinking
of the data. The visualization of the model is subse-
quently discussed in section 5, while the work is val-
idated in section 6. Finally, section 7 concludes the
paper.
2 DATA SOURCE
CHARACTERIZATION AND
SELECTION
Central to the development of a model is the data that
eventually will be represented in the model and thus
needs to be utilized for building and populating the
model. With the research question in mind, the iden-
tification, and subsequent selection, of data sources
that provide disease related information, pertaining
to, for example, symptoms, inheritability, and genet-
ics of a disease, thus are the first key steps in the
development process of the disease related informa-
tion model. A search for disease related information
results in a wide variety of structured (i.e standard-
ized terminologies or vocabularies, ontologies, and
databases) and unstructured (e.g. websites (U.S. Na-
tional Library of Medicine, 2015c; WebMD, LLC,
2015)) data sources. Data from unstructured sources
requires conversion to a structured format, for exam-
ple using Natural Language Processing (NLP) tech-
niques, and thus cannot be directly incorporated into
the disease related information model. As a result
we have decided to only incorporate structured data
sources. It is essential to designate a primary data
source for the development of the disease model as the
available structured data sources for disease related
information overlap in terms of covering the same in-
formation in different formats and presentations.
A disease model that is represented in a network-
like format consists of two components, namely con-
cepts and relationships among these concepts. Con-
cepts can be sourced from standardized terminolo-
gies, or from ontologies. Some well-known termi-
nologies in the biomedical field are the International
Classification of Diseases (ICD) (World Health Orga-
nization, 2015), Medical Subjects Headings (MeSH)
(U.S. National Library of Medicine, 2015a), and Sys-
tematized Nomenclature of Medicine Clinical Terms
(SNOMED CT) (International Health Terminology
Standards Development Organisation, 2015), whereas
the National Cancer Institute Thesaurus (NCIt) (U.S.
National Cancer Institute, 2015b), the Disease On-
tology (Institute for Genome Sciences - University
of Maryland School of Medicine, 2015), and the
Gene Ontology (Ashburner et al., 2000) are among
the frequently used ontologies within the biomedi-
cal field. Instead of sourcing concepts from one or
multiple individual terminologies, one can source the
concepts from the Unified Medical Language Sys-
tem (UMLS) Metathesaurus (U.S. National Library
of Medicine, 2015e) or the National Cancer Institute
Metathesaurus (NCIm) (U.S. National Cancer Insti-
tute, 2015a), both of which integrate, among many
others, the aforementioned sources into a single ter-
minology. Using these metathesauri provides the op-
portunity of broadening the scope of the concepts that
are covered and, as such, expanding the knowledge
base of the model by using concepts represented in
the majority of separate terminologies. Therefore, the
use of either the UMLS or NCIm to define concepts
in the model is preferred over the use of separate ter-
minologies.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
52

Figure 1: System Architecture of BioMed Xplorer.
Relationships, on the other hand, can also be
sourced from the UMLS and NCIm. More extensive
relationships, however, can be obtained from the On-
line Mendelian Inheritance in Man (OMIM) database
(Johns Hopkins University, 2015), MalaCards (Weiz-
mann Institute of Science, 2015), or SemMedDB
(Kilicoglu et al., 2012). Considering the overarch-
ing aim of this research in aiding (bio)medical re-
searchers in their knowledge explorations efforts, and
due to the fact that (bio)medical knowledge originat-
ing from peer-reviewed literature is considered trust-
worthy and rich, relationships directly derived from
(bio)medical literature are selected as the primary re-
lationships in the model. To this end, SemMedDB is
thus selected as the primary source, presenting dis-
ease related information, for incorporation into the
developed model. This choice is further motivated
by the fact that SemMedDB is considerably larger
(containing over 70 million statements) than the other
identified sources containing disease related informa-
tion. Finally, the broad scope, covering terms across
the entire biomedical domain, also played a role in the
choice for SemMedDB.
3 DATA PREPROCESSING
Due to the large amounts of heterogeneous and dy-
namic information that is nowadays available across
a multitude of sources, relational databases are con-
sidered to be less than ideal for storing and instanti-
ating knowledge representations of information with
such nature (Hendler, 2014). Linked data, on the
other hand, provides a promising solution to this issue
as it is able to cope with such large amounts of dy-
namic and heterogeneous information (Berners-Lee
et al., 2001). To this end we therefore aim to de-
velop our model using Semantic Web technologies.
According to (Berners-Lee et al., 2001; Antoniou and
Van Harmelen, 2004) the Semantic Web consists of
three main components, being i) labeled graphs that
encode meaning by representing concepts and the re-
lations among them, and are usually expressed as
(subject-predicate-object) triples in RDF, ii) Uniform
Resource Identifiers (URIs) to uniquely identify the
items in the datasets as well as to assert meaning,
which is reflected in the design of RDF, and iii) on-
tologies to formally define the relations that can ex-
ist among data items. In order to develop our model
using the Semantic Web, the existence of these three
components needs to be ensured. Processing the data
in SemMedDB such that these three components ex-
ist, is therefore the main aim of the preprocessing
stage.
3.1 Ontology Design
In order to be able to generate labeled graphs from a
relational database, such as SemMedDB, and to en-
sure the use of URIs, an ontology needs to be devel-
oped that represents the desired data structure of these
graphs. This ontology should define the data items,
as well as the relations among them, that are aimed to
be represented. Considering that the planned model
should represent the statements, and their provenance
data, in SemMedDB as a RDF graph, it is key for the
ontology to closely resemble SemMedDB’s database
design. Prior work has been conducted in this area by
(Tao et al., 2012). In their work, (Tao et al., 2012)
aimed to optimize the organization and representa-
tion of Semantic MEDLINE data (SemMedDB) for
translational science studies by reducing redundancy
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data
53

through the application of Semantic Web technolo-
gies. This is achieved by representing the concepts
and associations in SemMedDB as RDF. Despite suc-
cessfully decreasing the redundancy of the informa-
tion in SemMedDB, two shortcomings can be identi-
fied in the ontology that was developed by (Tao et al.,
2012). First of all, the ontology represents a limited
amount of information compared to the information
that is available in SemMedDB. This, in turn, impedes
the ability to incorporate external resources into the
model since among the information from SemMedDB
that is omitted are unique identifiers that are required
to retrieve the appropriate entities from these exter-
nal sources. The second shortcoming is the lack of
reuse of terms defined in existing vocabularies, which
is one of the founding principles of the Semantic Web
(Shadbolt et al., 2006). In (Tao et al., 2012), the devel-
oped ontology defines all terms used, whereas equiv-
alent classes might already exist in other vocabularies
in the Web of Data. Such reuse would facilitate the
linking of data to a Web of Data, which is an overar-
ching goal of the Semantic Web (Berners-Lee et al.,
2001).
Despite the limitations of the ontology developed
in (Tao et al., 2012), this ontology is considered as a
starting point as well as an opportunity to improve on
and extend upon. To this extent, the BioMed Xplorer
Ontology is developed that addresses the identified
shortcomings by representing most of the information
contained within SemMedDB, as well as by reusing
as much terms from existing vocabularies or ontolo-
gies as possible. The BioMed Xplorer Ontology is de-
veloped in the Web Ontology Language (OWL2) and
is published on a Persistent Uniform Resource Loca-
tor (PURL) (Weibel et al., 1996) domain
1
. Such a lo-
cator allows the underlying Web address of a resource
to change while not affecting the availability of the
systems that depend on this resource. The BioMed
Xplorer Ontology is shown in Figure 2.
RDF Reification. Considering that the provenance
data in SemMedDB applies to statements as a whole,
reification is necessary in order to represent this
provenance data in the ontology. (Tao et al., 2012)
also recognized this need, however, they did not use
the RDF Reification vocabulary as outlined in (World
Wide Web Consortium et al., 2014). The BioMed
Xplorer Ontology on the other hand implements the
RDF reification vocabulary.
As the statements contained in SemMedDB relate
two UMLS concepts to each other, both the subject
and object of an rdf:Statement instance are modelled
1
http://purl.org/net/fcnmed
as instances of a Concept class. The concepts are
related to each other through one, of 58, relation-
ships that are identified by SemRep (U.S. National
Library of Medicine, 2015d). The predicate of an
rdf:Statement instance therefore is modelled as one
of 58 instances of the Relation class. This set of re-
lationships consists of two disjunctive subsets, with
one subset containing 31 relationships derived from
the UMLS Semantic Network, such as ”causes”, and
the other subset containing the remaining 27 relation-
ships, which are negated versions of the relationships
in the first subset, such as ”neg causes”, referring
to ”does not causes” (Kilicoglu et al., 2012). Rela-
tionships belonging to the negated subset are prefixed
with ”NEG”, whereas all other relationships are con-
sidered to belong to the subset of affirmed relation-
ships. These two subsets of relations are represented
in the BioMed Xplorer Ontology as two subclasses of
the Relation class, being the AffirmedRelation and the
NegatedRelation classes respectively.
The provenance data in SemMedDB includes both
the sentences from which a statement is derived, as
well as the publications in which these sentences oc-
cur. Reification of the statements enables the asser-
tion of this provenance data to their respective state-
ments. To this end, sentences are represented as in-
stances of the Sentence class, which are related to the
rdf:Statement class through a derivedFrom property.
The articles in which these statements and sentences
are contained, are represented as instances of an Arti-
cles class, which are related to the rdf:Statement class
through a source property. Furthermore, sentences are
related to articles through the partOf property, indi-
cating that a sentence is part of an academic article.
In addition to the object properties, relating classes
to each other, discussed in this section, a number
of datatype properties, associating data values (such
as identifiers) to classes, are asserted to each of the
classes in the BioMed Xplorer Ontology as well. Col-
lectively, these properties aim to represent as much
information from SemMedDB in the ontology as pos-
sible.
Vocabulary Reuse. The BioMed Xplorer Ontology
aims to reuse as much existing classes and proper-
ties as possible. To this extent all elements of the
ontology, which include the classes and both the ob-
ject and datatype properties, except elements from the
RDF or RDFS namespaces, have been checked for
the presence of already defined equivalent concepts
or properties in existing vocabularies. This has been
accomplished by making use of the online RDF vo-
cabulary search and lookup tool vocab.cc (Institute
of Applied Informatics and Formal Description Meth-
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
54

Figure 2: BioMed Xplorer Ontology, only showing the object properties.
ods - Karlsruhe Research Institute, 2015) that allows
one to enter any term, and returns any classes and
properties that (partially) match the term. In gen-
eral the highest ranked term that corresponds to the
role of the term in the BioMed Xplorer Ontology
(e.g. class or property) is selected for reuse in the
BioMed Xplorer. In the end, the search for existing
terms lead to the incorporation of terms from three
existing vocabularies, being i) the Bibliographic On-
tology (D’Arcus and Giasson, 2015), ii) the Dublin
Core Metadata Terms (Dublin Core Metadata Initia-
tive (DCMI), 2015), and iii) the Simple Knowledge
Organization System (World Wide Web Consortium,
2015).
4 DATA INTERLINKING &
FUSION
The ontology developed in section 3.1 defines the de-
sired data structure for the developed model. Generat-
ing the labeled graphs from the SQL in SemMedDB,
however, requires a mapping that specifies how the
data in the database is matched and converted to
the appropriate class instances, properties, and prop-
erty values specified in the ontology. Such a map-
ping can be developed using D2RQ, a declarative
language for describing mappings between relational
databases, RDF(S), and OWL ontologies (Bizer and
Seaborne, 2004). The developed D2RQ mapping files
have been made available online
2
. A mapping file en-
ables RDF applications to access relational databases
as virtual RDF graphs through the companion tool
D2R Server (Bizer and Cyganiak, 2006). These vir-
tual RDF graphs can subsequently be queried using
2
The mapping files are available online from:
https://goo.gl/1yD0WO
the SPARQL protocol, with the D2RQ mapping trans-
lating the SPARQL queries to SQL queries, and trans-
lating the query results back to RDF. Both D2RQ
and D2R are jointly available in the D2RQ Platform
(Bizer and Seaborne, 2004). With the developed map-
ping file, the data in SemMedDB can be interlinked
as RDF triples according to the specified ontology, as
such surfacing and populating the actual disease re-
lated information model. Furthermore, the combina-
tion of the ontology and the use of RDF ensures the
ability to link to the data in the information model
from external datasets, through the URIs assigned to
instances and properties.
In order to achieve complete data fusion with ex-
ternal sources, as such creating a truly Linked Data
model, the data in the disease related information
model should be linked to related entities or instances
in external (RDF) data sources. This can be achieved
by setting RDF links between the data in the model
and these external data sources (Berners-Lee et al.,
2009). One common way of setting such links be-
tween data sets is through the owl:sameAs property,
which indicates that two linked individuals refer to
the same thing (Dean et al., 2004). Establishing these
links subsequently enables the incorporation of data
from the external data sources into the disease related
information model. Key to this data fusion process is
the identification of external data sources containing
instances that are equivalent to the instances in the
developed model. The search for these data sources
containing equivalent instances has been facilitated
by searching the Linked Open Data cloud
3
for unique
standardized instance identifiers. Among these iden-
tifiers in SemMedDB are the UMLS Concept Unique
Identifier (CUI), the Entrez-Gene ID, and the OMIM
identifier for concepts, as well as the PubMed Iden-
tifier (PMID) for publications. The search of the
3
For details see http://lod-cloud.net/
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data
55

Linked Open Data cloud for (bio)medical RDF data
sources that represent either (bio)medical concepts,
identified by one of the aforementioned identifiers,
or publications, identified by the PubMed identifier,
returned two main external data sources that could
be fused with the data in SemMedDB: Linked Life
Data (Momtchev et al., 2009), and Bio2RDF (Belleau
et al., 2008).
5 BIOMED XPLORER UI
Assisting (bio)medical researchers in their knowledge
exploration efforts can be achieved by enabling them
to intuitively explore the body of (bio)medical knowl-
edge. To this end it is therefore imperative to visualize
the developed disease related information graph, rep-
resenting this body of knowledge, that incorporates
other disease related information gathered and aggre-
gated from disperse sources across the Web. With
this in mind three key requirements for the BioMed
Xplorer UI can be imagined, being that it should:
i) be usable and intuitive (e.g. supported by an ap-
propriate visualization paradigm), ii) concisely repre-
sent provenance data (e.g. the publications as well
as sentences from which statements are derived), and
iii) represent information from multiple sources (e.g
concept summaries and definitions). Based on these
identified requirements, BioMed Xplorer has been
developed and made available
4
on the Web. The
BioMed Xplorer UI supports the visualization of the
information and has been developed in JavaScript in
combination with the d3.js
5
and jQuery
6
libraries.
The data visualized by the BioMed Xplorer UI
is obtained from BioMed Xplorer’s back-end, which
consists of a Virtuoso triple store containing the dis-
ease related information model in RDF. This triple
store provides a built-in SPARQL endpoint that can be
queried by BioMed Xplorer using the SPARQL (Har-
ris et al., 2013) protocol. Efficient query handling has
been achieved by the development of a caching mech-
anism.
The developed user interface has three key fea-
tures being: i) a graph-based visualization, ii) the ex-
ploration of concept information, and iii) the explo-
ration and assessment of relationships. Each of these
three features will be briefly discussed in the remain-
der of this section.
4
BioMed Xplorer is available http://goo.gl/qeuW5k
(best viewed in Firefox).
5
For details see http://d3js.org/
6
For details see http://jquery.com/
Graph Visualization. The BioMed Xplorer UI em-
ploys a graph-based visualization of (bio)medical
knowledge as shown in Figure 3. Exploration and
traversal of the knowledge graph is supported through
the expansion of concepts (by double clicking on con-
cepts) and collapsing of concepts (by right clicking on
concepts). Additionally, panning (by click and drag)
and zooming (by scrolling) is supported as well.
Exploring Concept Information. Within the
BioMed Xplorer UI concept information can be
explored through concept summaries, which can
be opened by clicking on concepts, and concept
overviews (as shown in Figure 4), which can be
opened by choosing to show details in a concept
summary. Concept information includes a wide range
of information available from within the model as
well as from external sources, such as Linked Life
Data and Bio2RDF.
Exploring and Assessing Relationships. Relation-
ships between concepts can be explored in the
BioMed Xplorer UI through statement summaries,
which can be opened by clicking on an edge, and
statement overview, which can be opened by choos-
ing to show details in a relationship summary. Within
statement overviews, a wide range of statement infor-
mation is available, as is shown in Figure 5. Among
the available information is: the complete statement,
two aggregates of the available provenance data, a
brief overview of the source and target concepts of
the relationship, the sentences from which the state-
ment is derived at a publication level, as well as the
details of the publications.
6 VALIDATION
Keeping the key roles of both the BioMed Xplorer
Ontology, as the the foundation for the knowledge
base, and the BioMed Xplorer UI, as the visualization
of this knowledge base in mind, the validation of these
two outcomes is imperative. This validation aims
to assess whether the proposed solutions successfully
address the already identified gap as well as how the
proposed solutions measure against existing work. To
this end, a two folded validation of both the ontology
and the visualization has been conducted. In this re-
gard a comparison to prior work has been conducted
first, the results of which are shown in Tables 1 and
2 respectively. Secondly, an evaluation has been per-
formed by 6 experts in the field. Results of this expert
evaluation showed that both the BioMed Xplorer On-
tology and the BioMed Xplorer UI successfully sat-
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
56
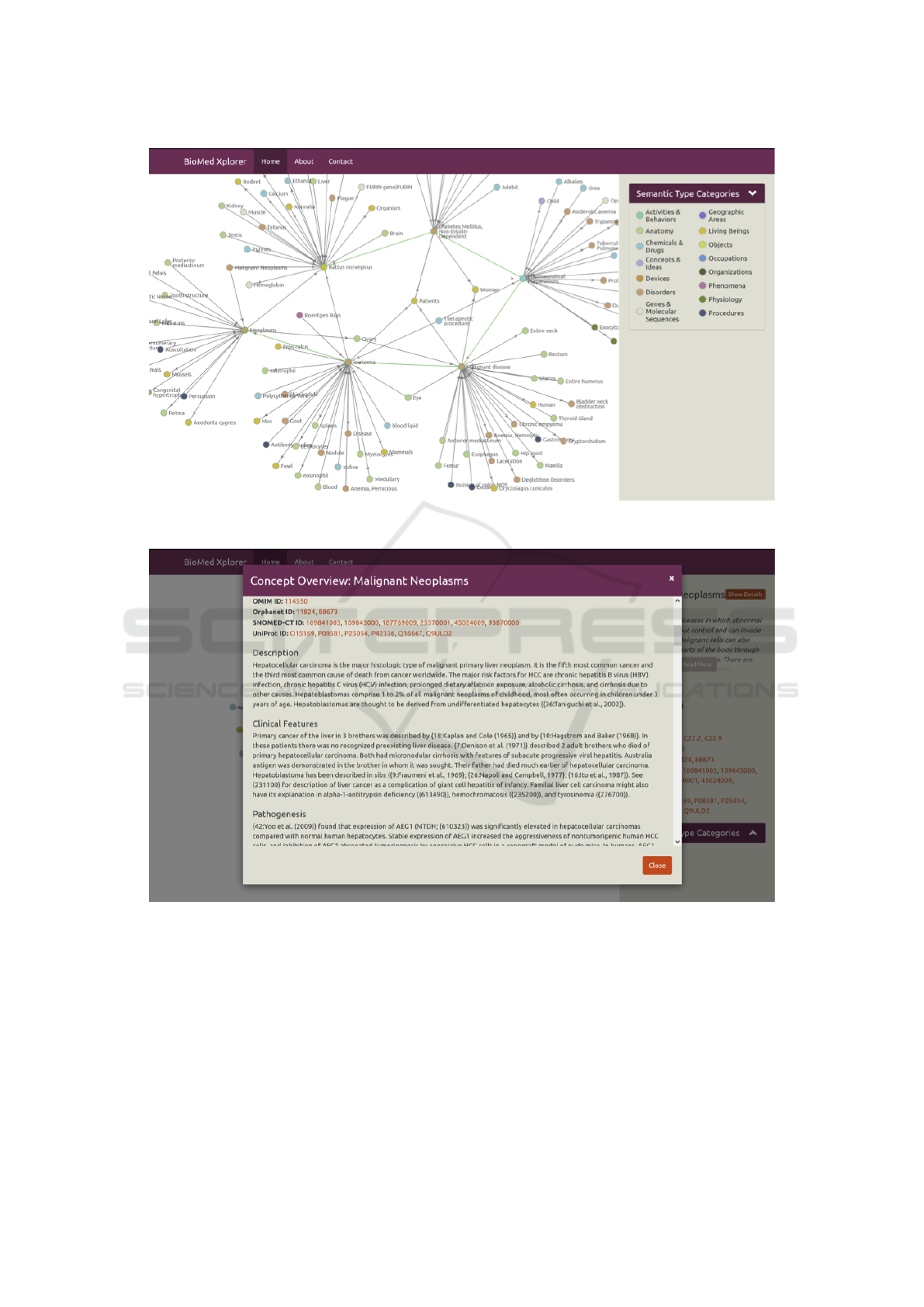
Figure 3: A screenshot from BioMed Xplorer UI and its graph-based visualization of (bio)medical knowledge, representing
(bio)medical concepts as nodes and their interrelationships as edges.
Figure 4: BioMed Xplorer UI’s Concept Overview for ”Malignant Neoplasms”.
isfy the identified requirements, with average grades
of a 7.8 and 7.6 out of 10 respectively. Details of the
expert evaluation of both the BioMed Xplorer Ontol-
ogy and the BioMed Xplorer UI are provided in Ta-
bles 3 and 4.
Comparison to Related Work. There are five main
knowledge representation and visualization tools
identified that attempt to address similar challenges
associated with exploring the body of (bio)medical
knowledge through the representation and visualiza-
tion of the knowledge contained within scientific pub-
lications. Among these tools are: AliBaba (Plake
et al., 2006), EBIMed (Rebholz-Schuhmann et al.,
2007), PGviewer (Tao et al., 2005), Semantic MED-
LINE (Kilicoglu et al., 2008), and the Semantic Nav-
igator (Bodenreider, 2000). Due to the close corre-
spondence between the aims of these tools and the
aims of our research, these five tools are considered
as the base for comparison to BioMed Xplorer. A
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data
57

Figure 5: BioMed Xplorer UI’s Statement Overview for ”Malignant Neoplasms” part of ”Rattus Norvegicus”.
Table 2: Comparison of BioMed Xplorer UI with five knowledge visualization tools developed in prior work.
Characteristic AliBaba EBIMed PG-
viewer
Semantic
MEDLINE
Semantic
Navigator
BioMed
Xplorer
Scope Limited Limited Limited Biomedical Biomedical Biomedical
Available No No No Yes Yes Yes
Visualization paradigm Graph Tabular Tree Graph Graph Graph
Concept categorization Yes Yes Yes Yes No Yes
Incorporation of links to external
sources
Yes Yes No Yes No Yes
Incorporation of data from external
sources
Yes Yes Yes No No Yes
Presentation of concept related infor-
mation
Yes No Yes Yes No Yes
Incorporation of provenance data Yes Yes Yes Yes No Yes
Table 1: Comparison of BioMed Xplorer Ontology with the
ontology developed by(Tao et al., 2012)
Characteristic Tao et al.,
2012
BioMed
Xplorer
Ontology
RDF Reification No Yes
Vocabulary reuse No Yes
Links to external
data sources
No Yes
Number of related
data-items captured
4 17
Provenance data cap-
tured
Publications Publications
and sentences
comparison of the characteristics of these five selected
tools is provided in Table 2.
AliBaba acts as an interactive tool that graphi-
cally summarizes the associations between concepts
from a rather limited sub-domain of the (bio)medical
field, namely between cells, diseases, drugs, proteins,
species, and tissues. AliBaba extracts these concepts
and the associations between them from scientific
publications that match a PubMed query.
Semantic MEDLINE provides similar functional-
ity in a broader domain as it uses concepts in the
UMLS Metathesaurus as its base. These concepts,
and their relationships, are extracted, respectively
identified, from the complete MEDLINE database,
and, similar to AliBaba, subsequently presented as a
graph. The Semantic Navigator also employs a graph-
based format. In this tool, the graph is used to repre-
sent the semantic structure of the UMLS, and as such
enables users to visually explore the concepts in the
UMLS as well as their relationships.
Contrary to the graph-based format employed by
AliBaba, Semantic MEDLINE, the Semantic Naviga-
tor, and BioMed Xplorer for visualizing (bio)medical
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
58

Table 3: Frequency distribution of the five point Likert-
scale scores for evaluating the BioMed Xplorer Ontology.
A score of 1 indicates disagreement and 5 indicates agree-
ment.
Statement 1 2 3 4 5
The ontology is capable of
representing (statements of)
biomedical knowledge
0 0 1 4 1
The ontology models (state-
ments of) biomedical knowl-
edge appropriately
0 1 1 3 1
The ontology is capable of rep-
resenting the provenance data
associated with (statements of)
biomedical knowledge
0 0 2 2 2
The ontology models prove-
nance data associated with
(statements of) biomedical
knowledge appropriately
0 1 1 2 2
The ontology globally fits its
purpose
0 0 2 2 2
Table 4: Frequency distribution of the five point Likert-
scale scores for evaluating the BioMed Xplorer UI. A score
of 1 indicates disagreement and 5 indicates agreement.
Statement 1 2 3 4 5
The implemented function-
alities support and facilitate
the exploration of biomedical
knowledge
0 1 1 1 3
Color coding of the nodes is
helpful
0 0 1 2 3
The information in the sum-
maries has a clear structure
0 1 2 1 2
The information in the sum-
maries is relevant
0 1 1 3 1
The information in the ex-
tended details has a clear struc-
ture
0 1 1 3 1
The information in the ex-
tended details is relevant
0 0 1 2 3
The interface is well structured
/ organized
0 0 2 3 1
The graphical user interface
has an adequate look and feel
0 0 2 2 2
The tool behaves as expected 0 2 2 1 1
The visualization is intuitive in
its use
0 2 1 2 1
The visualization of informa-
tion is simple and smooth
0 1 0 3 2
The system globally fits its pur-
pose
0 2 0 1 3
knowledge, EBIMed and PGviewer make use of
two alternative visualization paradigms. On the one
hand, EBIMed identifies relationships between a set
of (bio)medical concepts extracted from publications
that match a MEDLINE query, and visualizes these
in a tabular format. The concepts represented in
EBIMed stem from the (bio)medical subdomain con-
sisting of proteins, Gene ontology annotations, drugs,
and species. On the other hand, PGviewer employs
tree visualization, with the purpose of clustering, in
order to present relationships from the genotype and
phenotype subdomain that are stored both in struc-
tured, such as MEDLINE, and (unstructured) textual
databases, such as the OMIM.
The five tools identified from prior work, in sum-
mary, thus have two main shortcomings. On the
one hand they focus on a particular subdomain of
the (bio)medical field (AliBaba, EBIMed, PGviewer)
and, as such, inhibit the exploration of the body
of (bio)medical knowledge. On the other hand
they employ an alternative visualization paradigm
(EBIMed and PGviewer) that is less focused on the
visual representation of knowledge. The BioMed
Xplorer UI overcomes these shortcomings, as such
improving over most tools developed in prior work,
through its broad scope, aiming to cover the complete
(bio)medical domain, and its graph-based visualiza-
tion. More specifically, BioMed Xplorer can be con-
sidered to be on par with Semantic MEDLINE and
the Semantic Navigator as these two tools both fo-
cus on the entire (bio)medical field as well as employ
a graph-based paradigm for visualizing (bio)medical
knowledge. The three aforementioned tools are fur-
thermore available on the Web, whereas AliBaba,
EBIMed, and PGviewer are no longer available. The
position of BioMed Xplorer is further reinforced by
the fact that it is the only tool that is based on RDF,
which improves its ability to handle large amounts
of heterogeneous data from disperse sources. Other
tools, on the other hand, are based on traditional re-
lational databases, as such inhibiting their ability to
incorporate data from additional external sources into
these tools.
In addition to representing (bio)medical knowl-
edge through statements that relate two (bio)medical
concepts to each other, the presentation of concepts
and statements related information is also of great im-
portance, as it provides background knowledge about
the concepts involved in the statements or about the
statements themselves. To this end, BioMed Xplorer
is on par with all of the other tools considering the
presentation of statement related information. This
information typically includes the complete statement
itself, including its source and target concepts, the
type of the statement, as well as the provenance data
associated with the statements in terms of the abstract
or sentences, and publications from which the state-
ments were derived. Such provenance data is pro-
vided by all the tools included in the comparison, ex-
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data
59

cept for the Semantic Navigator as this tool solely rep-
resents the relationships stored in the UMLS. Occa-
sionally, the statement related information might also
include aggregates of the provenance data, such as the
number of sentences and publications from which a
particular statement is derived, as is the case for Se-
mantic MEDLINE and BioMed Xplorer. Whereas the
BioMed Xplorer is on par with the other tools in re-
lation to the presentation of statement related infor-
mation and the incorporation of provenance data, it
in fact improves over these tools on the presentation
of concepts related information. The concepts related
information presented in BioMed Xplorer UI extends
well beyond the conventional information that is in-
corporated. While, tools such as EBIMed and the Se-
mantic Navigator do not present any of such concepts
related information at all, data items such as (seman-
tic) types, synonyms, and parts of publications that
mention the particular concept are presented by Al-
iBaba, PGviewer, and Semantic MEDLINE. BioMed
Xplorer extends this further through the incorporation
of a wide range of cross-identifiers of concepts, a def-
inition, and a range of data items pertaining to the
clinical features, diagnosis, inheritance, pathogenesis,
and genetics of a disease from OMIM, if available.
The presentation of this wide range of concept related
information in BioMed Xplorer is partially facilitated
through the incorporation of data from external (struc-
tured) data sources, including Linked Life Data and
Bio2RDF, which demonstrates its superiority com-
pared to the other tools developed in prior work.
Among these other tools, the incorporation of infor-
mation from such external sources is either largely
absent (such as in Semantic MEDLINE and Seman-
tic Navigator), or limited to the inclusion of informa-
tion from PubMed (such as in AliBaba, EBIMed, and
PGviewer). Links to external data sources, usually
in the form of cross-references to standardized termi-
nologies, on the other hand, are commonly used by
the tools developed in prior work, with only PGviewer
and the Semantic Network lacking such cross refer-
ences.
For the validation of the ontology, the ontology
developed by (Tao et al., 2012) is considered as the
base to which our developed ontology is compared.
A comparison of the characteristics of the two ontolo-
gies is provided in Table 1. As is clear from this table,
the ontology developed in this research improves the
ontology developed by (Tao et al., 2012) on a number
of aspects, which will be further discussed below, as
such contributing to the validation of the ontology de-
veloped in this research. As was discussed in section
3.1, reification has been applied in both ontologies to
allow triples to involve a particular (bio)medical state-
ment, as a whole, into another statement, and thus
enable meta-statements: statements about statements.
This can be achieved by treating a statement, relat-
ing two (bio)medical concepts to each other through
a relation, as a separate entity to which the subject,
the predicate, and the object of the original statement
are assigned using an object property. The ontology
developed by (Tao et al., 2012) performs this by mak-
ing use of the Association class in combination with
the hass name, has predicate, and haso name prop-
erties. BioMed Xplorer Ontology, on the other hand,
makes use of the official RDF reification vocabulary
that uses the rdf:Statement class in combination with
the rdf:subject, rdf:predicate, and rdf:object proper-
ties. Additionally, the use of this official RDF reifi-
cation vocabulary also contributes to the reuse of ex-
isting vocabularies, one of the key principles of Se-
mantic Web (Shadbolt et al., 2006). To further pro-
mote this base principle of the Semantic Web, the de-
veloped ontology, in addition to the use of the RDF
reification vocabulary, makes extensive use of exist-
ing classes and properties from other vocabularies.
This is a considerable improvement over the ontology
developed by (Tao et al., 2012), as the reuse of exist-
ing vocabularies, aside from the RDF vocabulary, is
not present in their ontology.
Since the purpose of the developed model is to en-
able researchers to explore the body of (bio)medical
knowledge as well as its (disease) related information,
the amount of information captured by the ontology
is of great importance. To this extent, the ontology
developed by (Tao et al., 2012) can be considered as
rather limited due to the fact that there is no direct
evidence of the incorporation of any (disease) related
information beyond the three datatype properties as-
signing names to concepts and relations, as well as
identifiers to publications. The ontology developed in
our research improves on this point by associating 17
data-type properties to the ontology classes that can
be used to capture a wide range of (disease) related
information. This is further facilitated by the incor-
poration of RDF links to the equivalent resources in
external data sources, including Linked Life Data and
Bio2RDF, which contain a wealth of (disease) related
information. No such links are incorporated in the on-
tology developed by (Tao et al., 2012).
Finally, the developed ontology extends the ontol-
ogy developed by (Tao et al., 2012) by incorporat-
ing the sentences from which the represented state-
ments are derived, in addition to those publications
from which these sentences are a part, as a compo-
nent of the provenance data that is associated to the
statements. The incorporation of these sentences pro-
vides additional value to the disease related informa-
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
60

tion model as it enables the presentation of the direct
source of a particular statement, as opposed to the pre-
sentation of solely the publication from which a state-
ment is derived.
7 CONCLUSION
Two common shortcomings among (bio)medical
knowledge discovery representation and visualization
tools are the scarcity of the information that is rep-
resented, usually coming from a single source, as
well as the lack of intuitiveness. To address this gap,
this research aimed at developing a dynamic model
representing (bio)medical knowledge, available from
disperse sources across the Web, as a network of
inter-related (bio)medical concepts, while incorporat-
ing Semantic Web technologies to deal with large
amounts of dynamic and heterogeneous information.
To achieve this goal, a five phase research ap-
proach has been followed, consisting of: 1) State of
the Art Assessment, 2) Data Source Characterization
and Selection, 3) Data Preprocessing and Ontology
Design, 4) Data Interlinking and Fusion with exter-
nal sources, and 5) Model Visualization. Comple-
tion of these phases resulted in the development of
the BioMed Xplorer Ontology, providing a founda-
tion of the knowledge base, and the BioMed Xplorer
UI, acting as a visualization of the knowledge base.
Future work will focus on implementing key in-
dicators, representing the importance of instances, to
more efficiently regulate which concepts and state-
ments are presented to the user. To this end, indica-
tors such as the degree of concepts, or number of sen-
tences or publications from which a statement is de-
rived, might be used. A second point of future work
will focus on extending BioMed Xplorer’s function-
ality with extensive filtering options, as such enabling
the user to view important, or less important, concepts
and statements based on key indicators.
ACKNOWLEDGEMENTS
This work was carried out on the Dutch national e-
infrastructure with the support of SURF Foundation
7
.
We also like to thank the School of Medicine at Dem-
ocritus University of Trace for helping with some re-
quirements identification and validation.
7
For details visit: https://www.surf.nl/en/services-and-
products/hpc-cloud/index.html
REFERENCES
Antoniou, G. and Van Harmelen, F. (2004). A semantic web
primer. MIT press.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D.,
Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K.,
Dwight, S. S., Eppig, J. T., et al. (2000). Gene ontol-
ogy: tool for the unification of biology. Nature genet-
ics, 25(1):25–29.
Belleau, F., Nolin, M.-A., Tourigny, N., Rigault, P., and
Morissette, J. (2008). Bio2rdf: towards a mashup to
build bioinformatics knowledge systems. Journal of
biomedical informatics, 41(5):706–716.
Berners-Lee, T., Bizer, C., and Heath, T. (2009). Linked
data-the story so far. International Journal on Seman-
tic Web and Information Systems, 5(3):1–22.
Berners-Lee, T., Hendler, J., Lassila, O., et al. (2001). The
semantic web. Scientific american, 284(5):28–37.
Bizer, C. and Cyganiak, R. (2006). D2r server-publishing
relational databases on the semantic web. In Poster at
the 5th International Semantic Web Conference, pages
294–309.
Bizer, C. and Seaborne, A. (2004). D2rq-treating non-
rdf databases as virtual rdf graphs. In Proceed-
ings of the 3rd international semantic web conference
(ISWC2004), volume 2004. Citeseer Hiroshima.
Bodenreider, O. (2000). A semantic navigation tool for the
umls. In Proceedings of the AMIA Symposium, page
971. American Medical Informatics Association.
Cohen, A. M. and Hersh, W. R. (2005). A survey of current
work in biomedical text mining. Briefings in bioinfor-
matics, 6(1):57–71.
D’Arcus, B. and Giasson, F. (2015). Bibli-
ographic Ontology Specification (BIBO).
http://bibliontology.com/specification. [Online;
accessed August 28 2015].
Dean, M., Schreiber, G., Bechhofer, S., van Harmelen, F.,
Hendler, J., Horrocks, I., McGuinness, D. L., Patel-
Schneider, P. F., and Stein, L. A. (2004). Owl web
ontology language reference. W3C Recommendation
February, 10.
Dublin Core Metadata Initiative (DCMI) (2015). Dublin
Core (DC). http://dublincore.org/. [Online; accessed
August 28 2015].
Harris, S., Seaborne, A., and Prudhommeaux, E. (2013).
Sparql 1.1 query language. W3C Recommendation,
21.
Hendler, J. (2014). Data integration for heterogenous
datasets. Big data, 2(4):205–215.
Hunter, L. and Cohen, K. B. (2006). Biomedical language
processing: what’s beyond pubmed? Molecular cell,
21(5):589–594.
Institute for Genome Sciences - University of Maryland
School of Medicine (2015). Disease Ontology (DO).
http://disease-ontology.org/. [Online; accessed Au-
gust 28 2015].
Institute of Applied Informatics and Formal Description
Methods - Karlsruhe Research Institute (2015). Vo-
cab.cc. http://www.vocab.cc/. [Online; accessed Au-
gust 28 2015].
BioMed Xplorer - Exploring (Bio)Medical Knowledge using Linked Data
61

International Health Terminology Standards Development
Organisation (2015). Systematized Nomencla-
ture of Medicine Clinical Terms (SNOMED CT).
http://www.ihtsdo.org/snomed-ct. [Online; accessed
August 28 2015].
Johns Hopkins University (2015). Online Mendelian Inher-
itance in Man (OMIM). http://www.omim.org/. [On-
line; accessed August 28 2015].
Kilicoglu, H., Fiszman, M., Rodriguez, A., Shin, D., Rip-
ple, A., and Rindflesch, T. C. (2008). Semantic med-
line: a web application for managing the results of
pubmed searches. In Proceedings of the third interna-
tional symposium for semantic mining in biomedicine,
volume 2008, pages 69–76. Citeseer.
Kilicoglu, H., Shin, D., Fiszman, M., Rosemblat, G., and
Rindflesch, T. C. (2012). Semmeddb: a pubmed-
scale repository of biomedical semantic predications.
Bioinformatics, 28(23):3158–3160.
Lu, Z. (2011). Pubmed and beyond: a survey of web
tools for searching biomedical literature. Database,
2011:baq036.
Momtchev, V., Peychev, D., Primov, T., and Georgiev,
G. (2009). Expanding the pathway and interaction
knowledge in linked life data. Proc. of International
Semantic Web Challenge.
Plake, C., Schiemann, T., Pankalla, M., Hakenberg, J., and
Leser, U. (2006). Alibaba: Pubmed as a graph. Bioin-
formatics, 22(19):2444–2445.
Rebholz-Schuhmann, D., Kirsch, H., Arregui, M., Gaudan,
S., Riethoven, M., and Stoehr, P. (2007). Ebimedtext
crunching to gather facts for proteins from medline.
Bioinformatics, 23(2):e237–e244.
Shadbolt, N., Hall, W., and Berners-Lee, T. (2006). The
semantic web revisited. Intelligent Systems, IEEE,
21(3):96–101.
Tao, C., Zhang, Y., Jiang, G., Bouamrane, M.-M., and
Chute, C. G. (2012). Optimizing semantic medline
for translational science studies using semantic web
technologies. In Proceedings of the 2nd international
workshop on Managing interoperability and compleX-
ity in health systems, pages 53–58. ACM.
Tao, Y., Friedman, C., and Lussier, Y. A. (2005). Vi-
sualizing information across multidimensional post-
genomic structured and textual databases. Bioinfor-
matics, 21(8):1659–1667.
U.S. National Cancer Institute (2015a). NCI Metathesaurus
(NCIm). https://ncim.nci.nih.gov. [Online; accessed
August 28 2015].
U.S. National Cancer Institute (2015b). NCI Thesaurus
(NCIt). https://ncit.nci.nih.gov. [Online; accessed Au-
gust 28 2015].
U.S. National Library of Medicine (2015a).
Medical Subject Headings(MeSH).
http://www.nlm.nih.gov/mesh/meshhome.html.
[Online; accessed August 28 2015].
U.S. National Library of Medicine
(2015b). Medline fact sheet.
http://www.nlm.nih.gov/pubs/factsheets/medline.html.
[Online; accessed August 28 2015].
U.S. National Library of Medicine (2015c). Medline Plus.
http://www.nlm.nih.gov/medlineplus/. [Online; ac-
cessed August 28 2015].
U.S. National Library of Medicine (2015d). SemRep.
http://semrep.nlm.nih.gov. [Online; accessed August
28 2015].
U.S. National Library of Medicine (2015e). Uni-
fied Medical Language System (UMLS).
http://www.nlm.nih.gov/research/umls/. [Online;
accessed August 28 2015].
WebMD, LLC (2015). WebMD. http://www.webmd.com/.
[Online; accessed August 28 2015].
Weibel, S. L., Jul, E., and Shafer, K. E. (1996). PURLs:
Persistent uniform resource locators. OCLC Online
Computer Library Center.
Weizmann Institute of Science (2015). MalaCards.
http://www.malacards.org/. [Online; accessed August
28 2015].
World Health Organization (2015). Interna-
tional Classification of Diseases (ICD).
http://www.who.int/classifications/icd/en/. [On-
line; accessed August 28 2015].
World Wide Web Consortium (2015). Simple
Knowledge Organization System (SKOS).
http://www.w3.org/2004/02/skos/. [Online; ac-
cessed August 28 2015].
World Wide Web Consortium et al. (2014). Rdf 1.1 seman-
tics.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
62
