
A Structure based Approach for Accurate Prediction of Protein
Interactions Networks
Hafeez Ur Rehman
1
, Usman Zafar
1
, Alfredo Benso
2
and Naveed Islam
1,3
1
Department of Computer Science, National University of Computer & Emerging Sciences, Hayatabad, Peshawar, Pakistan.
2
Department of Control & Computer Engineering, Politecnico di Torino, I-10129, Torino, Italy.
3
Department of Computer Science, Islamia College University, Peshawar, Pakistan.
Keywords:
Protein Interactions, Protein Structure, 3D Templates, Protein Interaction Network, Protein Binding Sites.
Abstract:
In the recent days, extraordinary revolution in genome sequencing technologies have produced an overwhelm-
ing amount of genes that code for proteins, resulting in deluge of proteomics data. Since proteins are involved
in almost every biological activity, therefore due to this rapid uncovering of biological “facts”, the field of
System Biology now stands on the doorstep of considerable theoretical and practical advancements. Precise
understanding of proteins, specially their functional associations or interactions are inevitable to explicate how
complex biological processes occur at molecular level, as well as to understand how these processes are con-
trolled and modified in different disease states. In this paper, we present a novel protein structure based method
to precisely predict the interactions of two putative protein pairs. We also utilize the interspecies relationship
of proteins i.e., the sequence homology, which is crucial in cases of limited information from other sources
of biological data. We further enhance our model to account for protein binding sites by linking individual
residues in structural templates which bind to other residues. Finally, we evaluate our model by combining
different sources of information using Naive Bayes classification. The proposed model provides substantial
improvements in terms of accuracy, precision, recall when compared with previous approaches. We report an
accuracy of 90% when tested for a protein interaction network of yeast proteome.
1 INTRODUCTION
Proteins are the most essential macromolecules that
are involved in almost every biological activity. Our
knowledge of new proteins is increasing with a rapid
pace as next generation sequencing technologies are
uncovering new genomes. The knowledge of proteins
alone, is not sufficient since proteins rarely act in iso-
lation. The overall complexity of biological systems
at different levels primarily arise due to the combina-
torial interactions caused by the proteins in the cells.
One of the crucial step for understanding biological
cells as engineered systems is to map networks of
DNA-protein, RNA-protein and protein-protein inter-
actions (PPIs) of a species as completely and accu-
rately as possible. Precise knowledge of protein inter-
actions is also a precondition for fulfilling the promise
of preventive as well as personalized medicines that
which means more rational development of antibacte-
rial compounds, drugs, and vaccines.
The conventional wet lab experiments e.g., Yeast
two-Hybrid (Y2H) (Ito et al., 2001) screening,
Protein-fragment Complementation Assays (PCA)
(N. Pelletier et al., 1999), or co complex interaction
maps (that are attained by high-throughput Co-affinity
Purification followed by Mass Spectrometry (AP/MS)
to identify protein-protein (bait) interactions) (Rigaut
et al., 1999; A. Shoemaker and R. Panchenko, 2007a)
etc., are either slow, costly or prone to noise because
of the nature of these experiments. Moreover, the ex-
isting noise in protein interaction databases resulted
by these experiments, plus the deluge of protein data
produced by next generation sequencing technologies
motivates the need to make accurate computational
techniques that can precisely map the interactions of
proteins on genome wide scale.
Several computational techniques have been pro-
posed in the past that incorporate a wide variety
of data e.g., phylogenetic profiles, sequence homol-
ogy, and co-expression of genes etc., to accurately
infer genome-wide protein-protein interactions (A.
Shoemaker and R. Panchenko, 2007b; Salwinski and
Eisenberg, 2003). However, comparative studies ad-
vocate that the development of noise free protein in-
Rehman, H., Zafar, U., Benso, A. and Islam, N.
A Structure based Approach for Accurate Prediction of Protein Interactions Networks.
DOI: 10.5220/0005705002370244
In Proceedings of the 9th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2016) - Volume 3: BIOINFORMATICS, pages 237-244
ISBN: 978-989-758-170-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reser ved
237
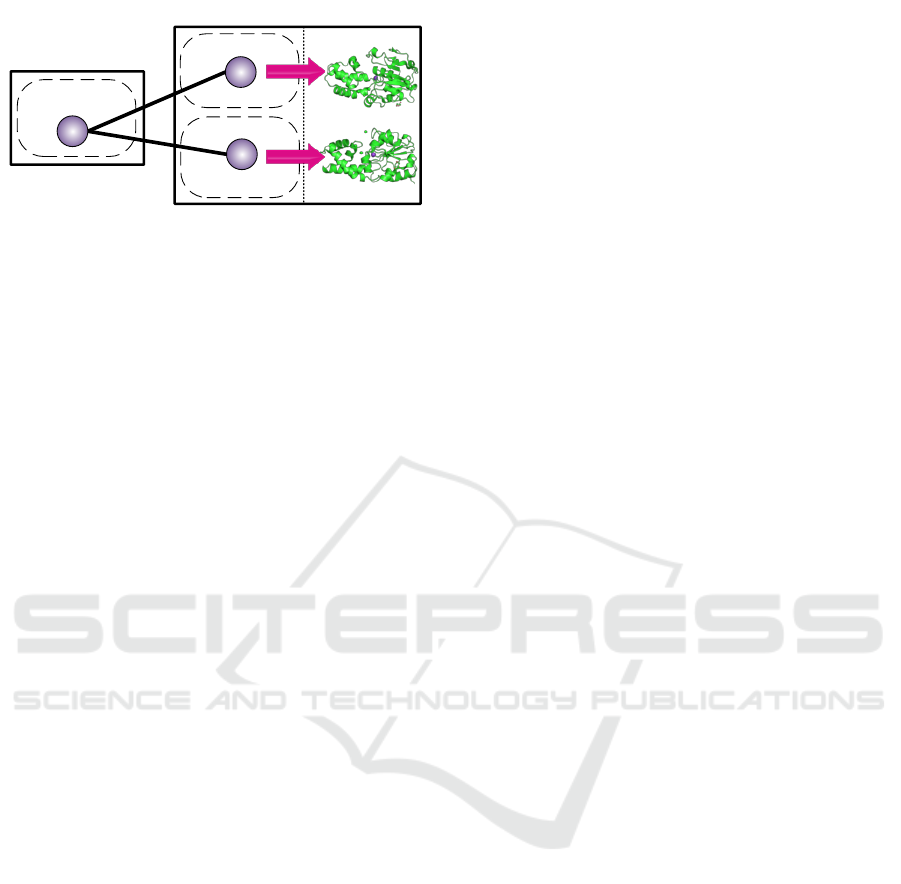
Homolog Species
Saccharomyces
Cerevisiae Species
Hypothetical Protein
YKL033W-A
HDHD1 Protein
CG15441 Protein
Drosophila
Melanogaster
Homo Sapiens
Known Structures
Figure 1: A hypothetical protein connected to structurally
known proteins using protein homology.
teraction repertoires of different genomes, is still in
its early stages (Braun and et al., 2009; Deane et al.,
2002). The most prominent computational methods
that produce high confidence interactions utilize pro-
tein’s structural information e.g., (C. Zhang et al.,
2010; Wass et al., 2011). But unfortunately, there is a
huge difference between the number of known protein
sequences and their relative known structures; even
for the well studied organism such as Saccharomyces
Cerevisiae, the known structural information is sparse
i.e. less than 10% proteins are with known structure
(Zhang et al., 2012). Moreover, the protein complex
information of known PPIs is even sparser.
Fortunately, homology models (see Figure 1) as
well as known protein complexes (across species) in
well-known databases e.g. PDB (Protein Data Bank)
(M. Berman et al., 2000), present the opportunity
to relate unknown structure sequences with known
structures using geometrical features of the individ-
ual templates. Approaches incorporating this type of
information have shown great success; in such cases
protein structure have multiple clues that associate
the geometric features of individual templates. How-
ever, these methods exhibit much less success on pro-
teins with inconsistent homolog templates (i.e. the
homolog templates whose geometrical features are
much variant; hence they result in effecting the over-
all accuracy of the prediction schemes).
In this paper we proposed and evaluated a novel
approach that combines heterogeneous structural in-
formation of proteins and determine their potency for
interaction in the form of a probability score. The fun-
damental conceptual innovation of our method is to
connect geometrical features of proteins with protein
binding sites and to enhance the algorithms power as
well as applicability for heterogeneous homolog tem-
plates. Our new approach relies on scores (features)
obtained by combining diverse sources of biological
information which includes: sequence similarity, pro-
tein homology, protein binding sites, and geometrical
features like, no of interacting residues, no of surface
residues etc. These scores are combined using Bayes
classifier and an overall confidence score is calculated
that determines the binding potential of two proteins
as interacting pairs.
The remaining part of the paper is organized as
follows: In Section 2, we first give an overview of
the closely related approaches used for the predic-
tion of protein-protein interactions; with the expla-
nation of why structure based approaches stand out
from other techniques. We then introduce, in Section
3, a heterogeneous information based Bayesian net-
work model that combines different types of informa-
tion (i.e., sequence similarity, protein homology, pro-
tein binding sites, and geometrical features) to predict
protein-protein interactions. Section 4 demonstrates
the effectiveness of the proposed model when applied
to cross validate, a subset of interacting as well as non
interacting proteins in the yeast network. We lastly
discuss the results of our scheme and also compare
the performance of most recent related state-of-the-art
structure based schemes with our scheme. In Section
5 we present conclusion of our study with possible
future considerations.
2 RELATED WORK
Protein-protein interactions are key to most of the bio-
logical processes. These interactions orchestracted by
molecular mechanisms that have yet not been clearly
understood. Understanding protein-protein interac-
tions would also provide us crucial clues about in-
terracellular signalling pathways. Numerous experi-
ments have been devised by researchers in the labs
including yeast two-hybrid systems, mass spectrom-
etry, protein microarrays and others. Unfortunately,
experimental techniques have not been able to charac-
terize the proteins to a great extent. Thus, our knowl-
edge of protein functions as well as their interactions
is very limited. This low contribution by experimental
techniques and lesser knowledge about protein inter-
actions is being complemented by the advancement of
computational methods.
Since, protein sequence is the most basic as well
as most easily available type of information about
proteins; therefore, many methods devised in the be-
ginning focused on use of sequence information to see
the mutual evolution of proteins. One such method,
focused on evolutionary information related to struc-
ture and function was proposed by (Valencia and Pa-
zos, 2003). This method constructs and utilized evo-
lutionary relationship among proteins to infer PPI as
such proteins co-evolve. Another approach was a
multiple classifier based system harnessing sequence
of proteins(F. Xia et al., 2010). They utilized two
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
238

classifiers rotation forest and autocorelation descrip-
tor. This group tested their system on Saccharomyces
cerevisiae and Helicobacter pylori data. Sequence
based approach for PPI prediction has been used by
another group but with slight variations. They pre-
dicted PPIs more precisely from sequence alignments
of proteins by using a Bayesian classifier (Burger and
V. Nimwegen, 2008). A similar and more recent set of
techniques utilized only sequence information for PPI
prediction e.g., (Shen et al., 2006; You et al., 2015).
Mathematical probabilistic models were adopted
by some researcher for the prediction of protein-
protein interactions. In one such case Probabilitic
analysis predicted nearly 40,000 interactions in hu-
mans (R. Rhodes et al., 2005). This probabilistic
model combined interaction data, functional anno-
tation data, protein domain data and genome-wide
gene expression data. Probabilistic models have also
provided a motivation for researches to model more
protein-protein interactions. A work was done us-
ing Generative Probabilistic Models with bi clique
perspective to model the interaction network of Sac-
charomyces cerevisiae(Schweiger et al., 2011). This
method concluded that nave unmodified DD (dupli-
cationdivergence) model is much more effective than
Preferential Attachment model at capturing key as-
pects of PPI prediction. Another work employed the
use of distant conservation of patterns in protein se-
quences, also called motifs and their structural rela-
tionships in proteins (Espadaler et al., 2005).
Most recent approaches that integrate structural
and non-structural type of information into compu-
tational models and use machine learning algorithms
e.g., Bayesian classifier or Support Vector Machines
etc., to infer interaction of putative proteins. One such
work is done in the recent past by (Zhang et al., 2012),
that utilizes structural as well as non-structural type of
features with a blend of Bayesian Classifier for pre-
diction of PPI on a genome wide scale. The authors of
this study presented their results for Saccharomyces
cerevisiae, and reported that structural features out-
perform non structural features with great margin in
terms of statistical performance measures i.e., pre-
cision, recall, accuracy, false positive rate etc., The
major contribution in this work was the use of struc-
tural features and evaluation of their impact on the
prediction accuracy. Thus structural information of
proteins plays a key role in deciphering the underly-
ing mechanism of protein interactions. Therefore, in
our work we also mainly integrate, structural infor-
mation of proteins along with protein binding sites to
predict their associations.
3 METHODS
In our work, we employ the idea of integrating hetero-
geneous biological information associated with two
queried proteins and determine their strength for in-
teraction, by combining this information in the form
of scores using Bayesian statistics. The distinctive-
ness of our technique comes from the fact that po-
tential interaction information e.g., protein binding
sites (which are strongly associated to molecular in-
teraction), can be combined with geometrical features
present in the structural templates of two interacting
proteins to decide if they interact or not. This com-
bination also increases the power of our algorithm to
include structural templates that are varied in geom-
etry but contain sites that can bind to other proteins.
Our proposed approach relies on scores (features) ob-
tained by combining diverse sources of biological in-
formation which includes: sequence similarity, pro-
tein homology, protein binding sites, and geometrical
features like, no of interacting residues, no of surface
residues etc.
The prediction of protein interactions is more
challenging for proteins which are not well annotated
or whose molecular details are limited. To enhance
the predictive power of our automated PPI predic-
tion algorithm, we combine very powerful associative
sources of information namely: protein homolog &
sequence similarity, as a baseline to capture proteins
which are most similar. This is particularly impor-
tant as each type of data typically captures distinct
aspects of associative activity. The overall process of
our technique for PPI prediction is divided into seven
steps (as shown in Figure 02):
Step 1: Selection of Homolog Sequences
To predict the interactions for sparsely annotated pro-
teins, the first useful type of information that can
associate them is the protein homology information.
Evolutionary relationships between species advocate
that the homolog (specifically orthologous) proteins
of different species, whose functions have been estab-
lished before speciation event and which share high
sequence similarity are likely to interact for similar
functional activities.
Two proteins are said to be homologous if they
share a common ancestor. To detect homology, se-
quence information is often used to deduce if proteins
are homologous or not. If two proteins share high se-
quence similarity i.e., above 25 % sequence similarity
(Benso et al., 2013; Mitrofanova et al., 2011; Benso
et al., 2012), they are very likely to be homologous
and have similar structures and in many cases part of
A Structure based Approach for Accurate Prediction of Protein Interactions Networks
239
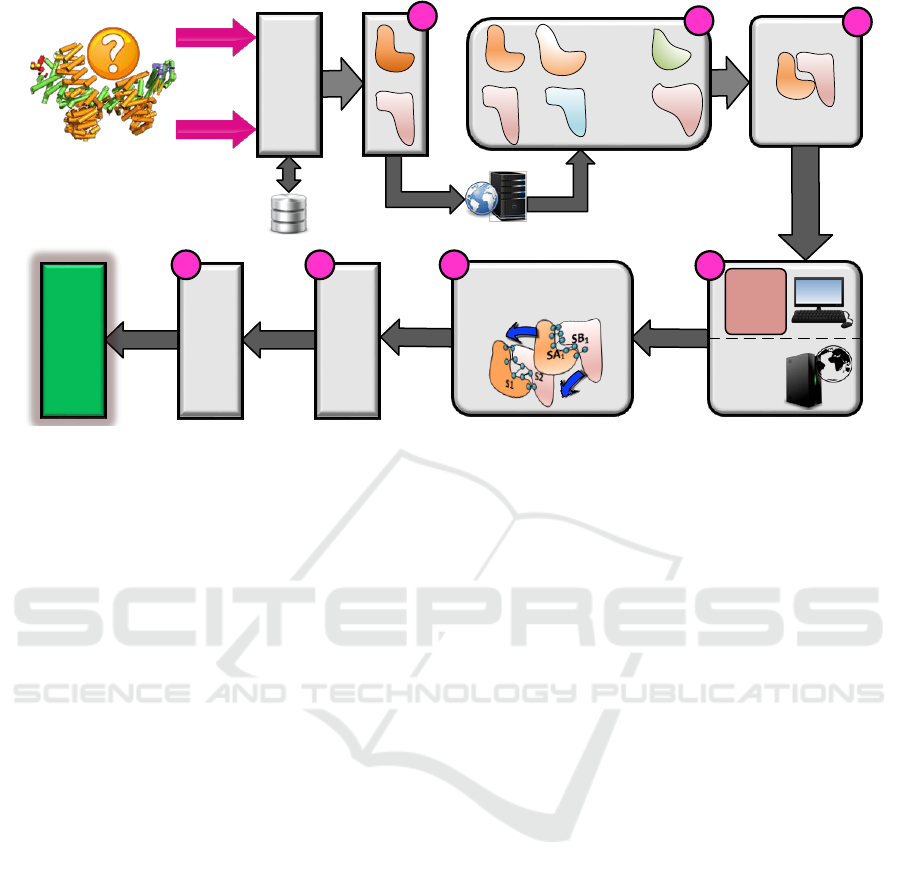
BLAST
S
1
S
2
Calculate
Scores
on all 100
Template
Model
Pairs
Bayesian
Classifier
Yes/No
Decision
SB
2
SB
j
SA
i
SA
1
SB
1
SA
2
S2N1
NR Database
VAST+ Server
Multiprot
Program to
Detect
Interacting
Residue Pairs in
the Structure
PDBe Binding
sites server
1
For i, j Templates
i x j Template Pairs in
Total
SA
1
Templates
2
3
4
56
Model by Superimposing
Templates with
Homologs
7
SB
1
Finds Structural
Neighbor Templates
of each Homolog
Structure
. . . . . . . .
. . . . . . . .
Query 1
Query 2
Homolog
PDB
Structures
Figure 2: The general shceme of heterogenous information integration for our PPI prediction algorithm.
the same molecular functional activity.
The input of our algorithm is a pair of proteins
(also called query proteins) say P
1
and P
2
(in our
implementation we used Uniprot IDs (The UniProt
Consortium, 2015)), whose interaction information
we want to find or predict. In the first step, since
these proteins can possibly be sparsely annotated so,
we need to associate them through homology to other
proteins. To capture ortholog based homolog similar-
ity we run a single iteration BLAST (Altschul et al.,
1990) search for each query protein P
i
against the pro-
tein NR database with an E-value cutoff of 0.0001.
We selected protein structures (namely, S
1
and S
2
;
also called Model structures) that are highly similar
to our query, with the additional constraint that match-
ing PDB structures should have at least 90% or higher
sequence identity. It is pertinent to note that this sim-
ilarity is only in sequences not in structure. The ob-
tained structures as a result of sequence similarity are
then queried to PDB (M. Berman et al., 2000), to ob-
tain their structural details (i.e. atomic coordinates,
residues information etc.).
Step 2: Finding Structural Neighbors
In the second step, structural representatives of each
model structure i.e. S
1
and S
2
, were taken directly
by querying each model structure to VAST+ (Vector
Alignment Search Tool Plus) Server (Madej et al.,
2013). VAST+ is a tool designed by NCBI (Na-
tional Center for Biotechnology Information) and uti-
lizes Molecular Modeling Database (MMDB), for 3-
dimensional structures, with the need of finding those
structures that have similar macromolecular com-
plexes. The macromolecular similarities are evalu-
ated using purely molecule’s geometric criteria, with-
out considering sequence similarity, thus it is able to
identify even distant homologs structures. We queried
VAST+ with default parameters and with a thresh-
old of ten templates i.e. we select top ten neighbor-
ing templates of each model structure. The structural
neighbors are named as SA
i
for model template S
1
and SB
i
for model template S
2
, (where, i = 1,2.....10)
.
Step 3: Formation of Templates Pairs
At this stage, we have 10 structural neighbors for each
query protein P
i
. To check the overall binding poten-
tial for interaction, of individual template pairs, we
construct pairs of each structural neighbor SA
i
with
SB
i
(where i = 1,2.....10) i.e., SA
1
pairs with SB
1
,
SB
2
.... and so on up to SB
10
, likewise we repeat pair-
ing for SA
2
, SA
3
.... up to SA
10
. This step results in a
total of 100 template pairs.
Step 4: Identification of Interacting
Residues and Binding Sites
As a first step, to evaluate the propensity for inter-
action, of individual template pairs, we first identify
the # of interacting residues in the template pairs. For
this purpose, we use a tool called Multiprot in a proto-
col known as PRISM (PRotein Interactions by Struc-
tural Matching) (Tuncbag et al., 2011; Shatsky et al.,
2004) . The Multiprot rationale is based on the fact
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
240

that globally different protein structures can interact
via chains of architecturally similar residues called
motifs. Thus Multiprot predicts binding residues by
utilizing structural similarity as well as evolutionary
conservation of putative binding residue also called
hot spots. For each template pair Multiprot calculates
the # of interacting residues.
To further strengthen the PPI prediction of our
technique we also utilize the PDBeMotif (Golovin
and Henrick, 2008). PDBeMotif is an incredibly fast
and powerful search tool that facilitates the explo-
ration of binding sites of single proteins or classes of
proteins e.g., Pepsin, and locates the conserved struc-
tural features of individual residues both within the
same specie as well as in different species. We em-
ploy PDBeMotif to locate residues that are binding
sites in our template pairs.
Step 5: Modeling Structural Templates
using Homolog Pairs
In this step, we build an interaction model Mod
ij
by
superposing the template pairs SA
i
and SB
j
over the
model template S
1
and S
2
. Overall 100 models are
built for (10x10) template pairs. Each model Mod
ij
is
used to calculate four structure based scores.
Step 6: Calculating Interaction Scores
from Interaction Models
From the 100 interaction models we prepared in the
previous step, we evaluate and combine associated in-
formation to calculate four scores for each interaction
model Mod
ij
. The scores are based on the criterion
that make use of interacting residues, binding sites
as well as sequence information. We name our first
score as ξ
(1)
Mod
i j
, where Mod
ij
denotes the interaction
model for which this score is calculated. ξ
(1)
Mod
i j
is
calculated by taking into account the number of in-
teracting residues in the template (calculated using
Multiprot) that are preserved in the homolog mod-
els S
1
and S
2
, i.e. both template and model share
those residue pairs. Templates have different varia-
tions in their amino acid sequence, this score captures
the strength of interaction model in terms of # of in-
teracting residues preserved, when compared with ho-
molog template pair.
The second score of our model is called ξ
(2)
Mod
i j
and is estimated by taking fraction of total interacting
residues preserved i.e., ξ
(1)
Mod
i j
, divided by the average
of total number of residues in both homolog templates
i.e., S
1
and S
2
, as shown in equation 1.
ξ
(2)
Mod
i j
=
ξ
(1)
Mod
i j
Average(S
1
,S
2
)
(1)
The third score ξ
(3)
Mod
i j
is the same as the first score,
with the additional check that the interacting residues,
both in template and model are also shared by the
binding sites retrieved using PDBeMotif service, and
is calculated as shown in the equation 2.
ξ
(3)
Mod
i j
=
h
ξ
(1)
Mod
i j
∩ Binding
−
Sites(Mod
i j
)
i
(2)
Lastly, the final score ξ
(4)
Mod
i j
, of our technique is
calculated by taking shared binding sites in the super-
imposed template and model pairs as shown in equa-
tion 3. ξ
(4)
Mod
i j
is the number of binding sites in the
template that align to the number of binding sites in
the model.
ξ
(4)
Mod
i j
=
Binding
−
Sites(S1, S2) ∩ Binding
−
Sites(Mod
i j
)
(3)
Step 7: PPI Prediction using Bayesian
Networks
Once all scores are calculated for hundred interaction
models, we then combine their effect into one score
by taking the mean and standard deviation of individ-
ual scores as shown in equation 4 and 5.
ϕ
(k)
=
∑
10
i=1
∑
10
j=1
ξ
(k)
Mod
i j
100
.. .For, k = {1, 2,3,4}
(4)
ϕ
(l)
=
v
u
u
t
∑
10
i=1
∑
10
j=1
ξ
(k)
Mod
i j
− ϕ
(k)
2
100
.. .For, k = {1, 2,3,4} and l = {5,6, 7,8} (5)
The Standard deviation of scores captures the fact
that, whether the templates that our method finds are
different from each other or not; because when dif-
ferences among homologs are spread out the standard
deviation will be high.
Lastly we use Bayesian classification to com-
bine the mean values as well as the standard de-
viations of our scores captured in eight variables
ϕ
(k)
, where k={1,2,...,8}. Let Pi and Pj be the
query proteins whose interaction we want to predict
A Structure based Approach for Accurate Prediction of Protein Interactions Networks
241

and ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
be the random vari-
able that capture different aspects of structural as-
sociation. The conditional probability that Pi and
Pj interact given the distribution of random variables
ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
is given by:
P(C
i j
= 1/ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
) =
P(ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
/C
i j
= 1).P(C
i j
= 1)
P(ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
)
=
Π
8
k=1
P(ϕ
(k)
/C
i j
= 1).P(C
i j
= 1)
6
Π
8
k=1
P(ϕ
(k)
/C
i j
= 1).P(C
i j
= 1)
+
Π
8
k=1
P(ϕ
(k)
/C
i j
= 0).P(C
i j
= 0)
(6)
Where P(C
i j
= 1) is the prior probability that
Pi and Pj interact, P(ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
)
is the probability that Pi and Pj has
ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
features and
P(ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
/C
i j
= 1) is the proba-
bility that Pi and Pj has ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
features given that Pi and Pj interact.
All feature values ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
are
normalized and we used binning of feature values
so that values of features lie in known ranges. As
many machine learning algorithms specially Bayes
classification produce better results when continuous
attributes are made discrete. Finally, we calculate
the value of P(C
i j
= 1/ϕ
(1)
,ϕ
(2)
,ϕ
(2)
,.........,ϕ
(8)
) for
each protein pair Pi and Pj.
4 EXPERIMENTAL SETUP AND
RESULTS
The integration technique described in the Methods
section is evaluated on the task of predicting protein-
protein interactions for an interaction network of Sac-
charomyces cerevisiae species proteins. We tested
our algorithm on a data set of Yeast species proteins
obtained from IntAct database [Results-01]. The al-
gorithm fuses probabilities derived from diverse data
sources including sequence similarity, protein homol-
ogy, protein binding sites used in combination with
other geometrical features. A well known power-
ful classification scheme i.e. Bayes classification,
was used to combine mutually independent features
(scores).
In this work, we present results of our scheme
for a portion of Saccharomyces cerevisiae species
interaction network. We chose the interaction net-
work of HSP75 YEAST protein (Uniprot ID: P11484)
for our experiment and tried to reproduce its inter-
action network using our proposed algorithm. The
HSP75 YEAST protein was chosen firstly because it
is involved in heterogeneous molecular activities and
secondly, because the interaction networks of this
protein contains many experimentally validated in-
teractions. Thus, to better evaluate the prediction
performance of our algorithm we chose this net-
work. The HSP75 YEAST is a fully reviewed protein
in UniProtKB/Swiss-Prot database (which is a high
quality manually curated, as well as non-redundant
protein sequence database).
The HSP75 YEAST protein’s interaction network
in IntAct database contain 4,449 interactions as of
August, 2015. The protein interaction network
databases contain false positive interactions that are a
bottleneck to predict the overall performance of an al-
gorithm as well as to judge the statistical significance
of experiments conducted. In order to deal with this
limitation, we filtered interaction network to include
interactions that are of high confidence with the crite-
ria that each interaction in the network must be sup-
ported by at least two experimental methods. After
filtering our network reduced to 1770 interactions.
We call these interactions as high confidence in-
teractions because each interaction is supported and
validated by at least two experimental methods. The
interaction network contains proteins from the same
(Saccharomyces cerevisiae) as well as other species
namely: Arabidopsis thaliana, Rattus norvegicus,
Arabidopsis thaliana, and Dictyostelium discoideum.
4.1 Performance Evaluation
For evaluating prediction performance we use cross
validation approach to estimate the prediction potency
of our proposed scheme i.e., for each protein pair Pi
and Pj in the interaction data set, we assumed the
interaction of Pi and Pj were unknown and then at-
tempted to predict the interaction by means of our
algorithm. Lastly, we compare the predicted inter-
actions with the true interaction set. For assessment
of our methodology, we computed performance mea-
sures, such as: precision, recall, accuracy and F1
which are estimated using the following formulas:
accuracy =
T P + T N
T P + FP + T N + FN
precision =
T P
T P + FP
recall =
T P
T P + FN
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
242

and
F1 =
2 ∗ precision ∗ recall
precision + recall
4.2 Cross Validation Analysis of
Prediction Accuracy
For the interaction network described earlier we first
attempted to predict protein-protein interactions by
10-fold cross validation. For each protein pair Pi and
Pj the probability that protein Pi interacts with Pj is
calculated using equation 6. Predicted protein interac-
tions having a probability estimate of greater than 0.5
were considered as positive interactions otherwise we
conclude that proteins don’t interact. By applying our
algorithm on high confidence interaction network re-
trieved from IntAct, we obtained an overall accuracy
of 90%, recall of 95.2%, precision of 94.11 and an F1
score of 94.49%.
4.3 Comparison with other Approaches
In this section, we broadly compare our method to the
most widely used group of techniques, such as Pre-
PPI algorithm proposed by Q. C. Zhang et al. (Zhang
et al., 2012), which combines structural as well as
non structural type of information to predict protein-
protein interactions. In such methodologies, inter-
actions among proteins are predicted by combining
structural clues with non structural clues using some
machine learning algorithm such as, Support Vector
Machines (SVM), Bayesian framework etc., which
consequently assign a probability score to a protein
pair of interest as positively or negatively interact-
ing. Fundamentals of Bayesian techniques are at the
heart of the overwhelming majority of methods cur-
rently used to combine heterogeneous sources infor-
mation for PPI prediction. Since this scheme (Zhang
et al., 2012), uses Bayesian technique as well as uti-
lizes structural information to predict PPI therefore,
we compare our algorithm against this computational
technique.
To obtain the most correct comparative results,
we use the same species proteins i.e., Saccharomyces
cerevisiae and compare results in a 10 fold cross-
validation setting. The results in figure 03 clearly sig-
nify that our method performs better than the Q. C.
Zhang’s Pre-PPI method (Zhang et al., 2012) across
all measures reported i.e., precision, recall, accuracy
and F1 scores. We observed that for almost the same
accuracy values, Pre-PPI method produced higher
number false positive as well as false negatives pre-
dictions, which resulted in lower values of precision
Acurracy
Recall
Precision
F1
Our Method
90
95.2
94.11
94.49
Pre-PPI Method
89
94
92
92
0
10
20
30
40
50
60
70
80
90
100
Percentage of values corresponding to each measure
Comparison with Pre-PPI
Figure 3: Comparison of Acurracy, Recall, Precision, and
F1 measure of proposed scheme with Pre-PPI scheme.
and recall, respectively. The improved performance
of our algorithm can be attributed to the most impor-
tant functional clue called protein binding sites, which
was further improved by combining with other struc-
tural information to precisely model the interaction
activity.
5 CONCLUSIONS
In this work, we presented a novel approach that uses
heterogeneous biological information associated with
two queried proteins and determine their strength for
interaction, by combining this information in the form
of scores using Bayesian statistics. The distinctive-
ness of our technique comes from the fact that po-
tential interaction information i.e., protein binding
sites, can be combined with other geometrical fea-
tures present in the structural templates of two inter-
acting proteins to decide if they interact or not. This
combination also increases the power of our algo-
rithm to include structural templates that are varied
in geometry but contain sites that can bind to other
proteins. The proposed model provides substantial
improvements in terms of accuracy, precision, recall
when compared with previous approaches. The pro-
posed scheme may additionally be used in combina-
tion with non structural features to enhance the pre-
diction confidence.
ACKNOWLEDGEMENTS
We would like to show our gratitude to Dr. Omar
Usman, Assistant Professor at National University of
Computer & Emerging Sciences for his worthy com-
ments that greatly improved the manuscript.
A Structure based Approach for Accurate Prediction of Protein Interactions Networks
243

REFERENCES
A. Shoemaker, B. and R. Panchenko, A. (2007a). De-
ciphering protein-protein interactions. part i. exper-
imental techniques and databases. PLOS Comput.
Biol., 3(3):e42.
A. Shoemaker, B. and R. Panchenko, A. (2007b). Deci-
phering protein-protein interactions. part ii. computa-
tional methods to predict protein and domain interac-
tion partners. PLOS Comput. Biol., 3(3):e43.
Altschul, S., Gish, W., Miller, Myers, E., and J. Lipman, D.
(1990). Basic local alignment search tool. Molecular
Biology, 215:403–410.
Benso, A., Di Carlo, S., Ur Rehman, H., Politano, G.,
Savino, A., and Suravajhala, P. (2012). Using gnome
wide data for protein function prediction by exploit-
ing gene ontology relationships. pages 497–502. IEEE
International Conference on Automation Quality and
Testing Robotics (AQTR)., IEEE.
Benso, A., Di Carlo, S., Ur Rehman, H., Politano, G.,
Savino, A., and Suravajhala, P. (2013). A combined
approach for genome wide protein function annota-
tion/prediction. PROTEOME SCIENCE, 11(S1):1–
12. ISSN: 1477-5956.
Braun, P. and et al. (2009). An experimentally derived con-
fidence score for binary protein-protein interactions.
Nature Methods, 6:91 to 97.
Burger, L. and V. Nimwegen, E. (2008). Accurate pre-
diction of protein protein interactions from sequence
alignments using a bayesian method. Mol Syst Biol,
4:165.
C. Zhang, Q., Petrey, D., Norel, R., and Honig, B. (2010).
Protein interface conservation across structure space.
Proc. Natl Acad. Sci. USA, 107:10896–10901.
Deane, C. M., Salwinski, L., Xenarios, I., and Eisenberg,
D. (2002). Protein interactions: two methods for as-
sessment of the reliability of high throughput observa-
tions. . Mol. Cell. Proteomics, 1:349 to 356.
Espadaler, J., Romero, O., M. Jackson, R., and et al. (2005).
Prediction of protein-protein interactions using distant
conservation of sequence patterns and structure rela-
tionships. Oxford Journals, Volume 21, Issue 16:3360
–3368.
F. Xia, J., Han, K., and S. Huang, D. (2010). Sequence-
based prediction of protein-protein interactions by
means of rotation forest and autocorrelation descrip-
tor. Protein Pept Lett, 17(1):137–45.
Golovin, A. and Henrick, K. (2008). Msdmotif: exploring
protein sites and motifs. BMC Bioinformatics, 9:1–11.
Springer-Verlag Berlin Heidelberg.
Ito, T., Chiba, T., Ozawa, R., and et al. (2001). A com-
prehensive analysis of protein protein interactions in
saccharomyces cerevisiae. Proc Natl Acad Sci USA,
98:4569–74.
M. Berman, H., Westbrook, J., Feng, Z., Gilliland, G.,
N. Bhat, T., Weissig, H., N. Shindyalov, I., and E.
Bourne, P. (2000). The protein data bank. Nucleic
Acids Research, 28:235–242.
Madej, T., J. Lanczycki, C., Zhang, D., A. Thiessen,
P., C. Geer, R., M. Bauer, A., and H. Bryant, S.
(2013). Mmdb and vast+: tracking structural simi-
larities between macromolecular complexes. Nucleic
Acids Res., 42:(D1): D297–D303. [PubMed PMID:
24319143].
Mitrofanova, A., Pavlovic, V., and Mishra, B. (2011). Pre-
diction of protein functions with gene ontology and in-
terspecies protein homology data. IEEE/ACM Trans-
actions on Computational Biology and Bioinformat-
ics, 8 no. 3:775–784.
N. Pelletier, J., Arndt, K., Pluckthun, A., and et al. (1999).
An in vivo library versus library selection of opti-
mized protein protein interactions. Nat Biotechnol,
17:683–90.
R. Rhodes, D., A. Tomlins, S., and Varambally, S. (2005).
Probabilistic model of the human protein-protein in-
teraction network. Nature Biotechnology, 23:951 –
959.
Rigaut, G., Shevchenko, A., Rutz, B., and et al. (1999). A
generic protein purification method for protein com-
plex characterization and proteome exploration. Nat
Biotechnol, 17:1030–32.
Salwinski, L. and Eisenberg, D. (2003). Computational
methods of analysis of protein protein interactions.
Curr. Opin. Struct. Biol., 13:377 to 382.
Schweiger, R., Linial, M., and Linial, N. (2011). Gener-
ative probabilistic models for protein-protein interac-
tion network the biclique perspective. Oxford Jour-
nals, Volume 27.
Shatsky, M., Nussinov, R., and J. Wolfson, H. (2004). A
method for simultaneous alignment of multiple pro-
tein structures. PROTEINS: Structure, Function, and
Bioinformatics, 56:143–156.
Shen, J., Zhang, J., Luo, X., Zhu, W., Yu, K., and et al.
(2006). Predicting protein-protein interactions based
only on sequences information. Proceedings of the
National Academy of Sciences, vol. 104:4337–4341.
The UniProt Consortium (2015). Uniprot: a hub for protein
information. Nucleic Acids Res. 43: D204-D212.
Tuncbag, N., Gursoy, A., Nussinov, R., and Keskin, O.
(2011). Predicting protein-protein interactions on a
proteome scale by matching evolutionary and struc-
tural similarities at interfaces using prism. Nature Pro-
tocols, 06 NO.09:1341–1354.
Valencia, A. and Pazos, F. (2003). Prediction of protein-
protein interactions from evolutionary information.
Methods Biochem Anal, 44:411–26.
Wass, M., Fuentes, G., Pons, C., Pazos, F., and Valencia,
A. (2011). Towards the prediction of protein interac-
tion partners using physical docking. Mol. Syst. Biol.,
7:469.
You, Z. H., Chan, K. C. C., and Hu, P. (2015). Predict-
ing protein-protein interactions from primary protein
sequences using a novel multi-scale local feature rep-
resentation scheme and the random forest. PLoS ONE,
10(5).
Zhang, Q. C., Petrey, D., and et al. (2012). Structure based
prediction of protein-protein interactions on a genome
wide scale. Nature, 490(7421):556 to 60.
BIOINFORMATICS 2016 - 7th International Conference on Bioinformatics Models, Methods and Algorithms
244
