
High-Throughput Sequencing Technology and Its Applications in
Human Disease
Shuyang Deng
1
1
College of Biotechnology Science and Engineering, Beijing University of Agriculture, Chang-ping District, Hui-longguan
Town,Beijing, 102206 China
512090957@qq.com
Keywords: High-Throughput Sequencing Technology; Disease; Exome Sequencing; Chip.
Abstract: The high-throughput sequencing technology (HTS), also known as next generation sequencing, refers to the
technological advances in DNA sequencing instrumentation that enable the generation of hundreds of
thousands to millions of sequence reads per run. The advances of high-throughput, low cost and short time-
consuming democratizes HTS and paves the way for the development of a large number of novel HTS
applications in basic science as well as in translational research areas, such as clinical diagnostics,
agrigenomics, and forensic science. In recent years, HTS has been widely applied in solving biological
problems, especially in human diseases field. In this review, we provide an overview of the evolution of
HTS and discuss three important sequencing strategies HTS adopted, Roche/454, Illumina, SOLiD. We also
take the example of exome sequencing and ChIP to summarize the application of HTS in human diseases.
1 INTRODUCTION
The human genome sequence has profoundly altered
our understanding of biology, human diversity, and
disease (Reuter, 2015). The human genome project,
however, required vast amounts of time and
resources and it is clear that faster, higher
throughput, and cheaper technologies. This
stimulates the development and commercialization
of high-throughput sequencing (HTS) technologies,
as opposed to the automated Sanger method, which
is considered a first-generation technology (Van Dijk,
2014).
Compared to first-generation Sanger sequencing
technology, the second-generation sequencing
technology not only maintains high accuracy, but
also dramatically increasing sequencing speed and
reduces sequencing costs. The Human Genome
Project lasted 13 years at a cost of nearly $ 3 billion,
and just created a single human genome map. At
present, the genome sequencing using a second-
generation sequencing technology costs only a few
thousand dollars, and the cost is still decreasing. In
the world’s top several genome research centers,
hundreds of people genome sequencing in a month
has been achieved. This new generation of high-
throughput analysis allows people to study the
disease with lower cost, more comprehensively. It
breaks down the limitations of previous fluxes on
disease research, which makes it possible to expand
all-round research on disease from genomic level,
transcriptome level, Proteomics level and other
aspects, shown by Fig. 1
Fig. 1. Disease research strategies of HTS.
The research and applications of HTS in the field
of life sciences and medical are becoming more and
more widespread. The following will introduce the
important roles of HTS in disease diagnosis and
prevention from three aspects of the prenatal care,
318
318
Deng S.
High-Throughput Sequencing Technology and Its Applications in Human Disease.
DOI: 10.5220/0006449703180324
In ISME 2016 - Information Science and Management Engineering IV (ISME 2016), pages 318-324
ISBN: 978-989-758-208-0
Copyright
c
2016 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tumor diagnosis and other major disease
pathogenesis.
HTS has been recognized in the field of
noninvasive prenatal care. Norton et al. (Norton,
2015) assigned pregnant women presenting for
aneuploidy screening at 10 to 14 weeks of gestation
to undergo both standard screening and cfDNA
testing. Of 18,955 women who were enrolled, results
from 15,841 were available for analysis. The AUC
for trisomy 21 was 0.999 for cfDNA testing and
0.958 for standard screening (P=0.001). Xu et al.
(Xu, 2015) carried out sequencing for single
blastomere cells and the family trio and further
developed the analysis pipeline, including recovery
of the missing alleles, removal of the majority of
errors, and phasing of the embryonic genome. The
final accuracy for homozygous and heterozygous
single-nucleotide polymorphisms reached 99.62%
and 98.39%, respectively. In addition, HTS can also
be applied to inherited heart diseases (Wilson, 2015),
cleft lip and palate (Wolf, 2015), phenylketonuria
(Gu, 2014) and so on, to prevent birth defects.
Currently, cancer is one of the major diseases
that endanger human health. As a complex disease,
its pathogenesis, typing and evolution are scientific
problems to be solved urgently. Besides, the
differences among individual cancer patients are
major challenge faced by the clinical treatment.
Using high-throughput DNA sequencing technology,
the comprehensive and systematic research on
cancer in the level of molecular biology can get a
large number of multi-dimensional tumor data set,
and extract tumor-associated genomic mutations or
modification information by scientific statistical
analysis. These will contribute to cancer prevention,
diagnosis, treatment, and overcome the road to lay
the cornerstone of the tumor for the human.
After describing the development of next-
generation sequencing in basic and clinical research,
Renkema et al. (Renkenma, 2014) suggested that
integrate data obtained using next-generation
sequencing with personalized medicine, including
use of high-throughput disease modelling as a tool to
support the clinical diagnosis of kidney diseases. To
develop an amplicon-based, next-generation
sequencing, mutation-detection assay for lung cancer
using the 454 GS Junior, Deeb et al. (Deeb, 2015)
designed fusion primers incorporating target
sequence, 454 adaptors, and multiplex identifiers to
generate 35 amplicons (median length 246 base
pairs) covering 8.9 kilobases of mutational hotspots
in AKT1, BRAF, EGFR, ERBB2, HRAS, KRAS,
NRAS, PIK3CA, and MAP2K1 genes and all exons
of the PTEN gene. In total, 25 point mutations and 4
insertions/deletions (indels) with a frequency of
5.5% to 93.1% mutant alleles were detected. Chung
et al. (Chuang, 2015) sought to investigate the
precise mutational landscape of four well-validated
Genetically engineered mouse models (GEMMs)
representing three types of cancers, non-small cell
lung cancer (NSCLC), pancreatic ductal
adenocarcinoma (PDAC) and melanoma. Thibodeau
et al. (Thibodeau, 2016) used next generation
sequencing to identify a pattern of genomic variation
associated with the development of brain metastases
in non-small cell lung cancer (NSCLC). While no
single variant was associated with brain metastasis,
this study implicated PI3K/AKT signaling and, in
particular, variants of TP53 as crucial for
determining the potential development of NSCLC
brain metastasis.
HTS has also been applied to great effect in the
field of other diseases. Information gained from
high-throughput DNA sequencing of
immunoglobulin genes (Ig-seq) can be applied to
detect B-cell malignancies with high sensitivity, to
discover antibodies specific for antigens of interest,
to guide vaccine development and to understand
autoimmunity (Georgiou, 2014). Redin et al. (Redin,
2014) reported the alternative strategy of targeted
high-throughput sequencing of 217 genes in which
mutations had been reported in patients with
intellectual disability or autism as the major clinical
concern. They analysed 106 patients with intellectual
disability of unknown aetiology following array-
CGH analysis and other genetic investigations. Saare
et al. (Saare, 2014) used a novel approach to
determine the endometriotic lesion-specific miRNAs
by high-throughput small RNA sequencing of paired
samples of peritoneal endometriotic lesions and
matched healthy surrounding tissues together with
eutopic endometria of the same patients. Results
indicated that only particular miRNAs with a
significantly higher expression in endometriotic cells
can be detected from lesion biopsies, and can serve
as diagnostic markers for endometriosis. Krauskopf
et al. (Krauskopf, 2015) focused on examining
global circulating miRNA profiles in serum samples
from subjects with liver injury caused by accidental
acetaminophen (APAP) overdose. Upon applying
next generation high-throughput sequencing of small
RNA libraries, they identified 36 miRNAs, including
3 novel miRNA-like small nuclear RNAs, which
were enriched in the serum of APAP overdosed
subjects. Comas et al. (Comas, 2016) described
current next generation sequencing approaches
applied to the Mycobacterium tuberculosis complex,
their contribution to the diagnostics and
High-Throughput Sequencing Technology and Its Applications in Human Disease
319
High-Throughput Sequencing Technology and Its Applications in Human Disease
319

epidemiology of the disease and the efforts that were
being undertaken to make the technology accessible
to public health and clinical microbiology
laboratories.
2 THE FUNDAMENTAL OF HTS
A. Roche/454
In the Roche/454 approach, the library fragments are
mixed with a population of agarose beads whose
surfaces carry oligonucleotides complementary to
the 454-specific adapter sequences on the fragment
library, so each bead is associated with a single
fragment (Mardis, 2008). Water micelles that also
contain PCR reactants, and thermal cycling
(emulsion PCR) of the micelles produces
approximately one million copies of each DNA
fragment on the surface of each bead. These
amplified single molecules are then sequenced en
masse. First the beads are arrayed into a picotiter
plate (PTP; a fused silica capillary structure) which
holds a single bead in each of several hundred
thousand single wells, which provides a fixed
location at which each sequencing reaction can be
monitored. Enzymecontaining beads that catalyze
the downstream pyrosequencing reaction steps are
then added to the PTP and the mixture is centrifuged
to surround the agarose beads. The PTP is seated
opposite a CCD camera that records the light emitted
at each bead. The first four nucleotides (TCGA) on
the adapter fragment adjacent to the sequencing
primer added in library construction correspond to
the sequential flow of nucleotides into the flow cell.
This strategy allows the 454 base-calling software to
calibrate the light emitted by a single nucleotide
incorporation.
B.
Illumina/Solexa
The Illumina system utilizes a sequencing-by-
synthesis approach in which all four nucleotides are
added simultaneously to the flow cell channels,
along with DNA polymerase, for incorporation into
the oligo-primed cluster fragments. Specifically, the
nucleotides carry a base-unique fluorescent label and
the 3’-OH group is chemically blocked such that
each incorporation is a unique event. An imaging
step follows each base incorporation step, during
which each flow cell lane is imaged in three 100-tile
segments by the instrument optics at a cluster density
per tile of 30,000. After each imaging step, the 3’
blocking group is chemically removed to prepare
each strand for the next incorporation by DNA
polymerase (Mardis, 2008).
C.
Solid
SOLiD systems isolate and amplify single DNA
molecules to construct a library for sequencing by a
process known as emulsion PCR (Tawfik, 1998).
Emulsification of an oil-water interface leads to the
formation of droplets, with each droplet, referred to
as a microreactor, containing a bead that is
covalently bound to a single DNA template. PCR
amplification is then performed across the surface of
the bead to generate clonally amplified fragments.
For SOLiD, after emulsion PCR, the 3’ ends of the
DNA template on the bead are modified to permit
chemical linkage to the surface of a glass slide.
When sequencing reagents containing DNA ligase
are flowed over the slide, a fluorescent signal is
generated that is captured by a CCD camera for base
calling. SOLiD sequencing is classified as
sequencing-by-ligation, because sequencing is
determined according to the selective mismatch
sensitivity of DNA ligase to fluorescently labeled
probes.
D.
Evolution of HTS Platform
Comparing with 454, Illumina and SOLiD
sequencing are more suitable, because of their higher
throughput. For this reason, transcriptome profiling
and ChIP-seq studies have mostly used Illumina or
SOLiD sequencing (Wang, 2009; Park, 2009). By
contrast, the reads generated by these technologies
are initially too short for de novo genome
assemblies. Thus, 454 is the preferred technology for
this type of application and enables exciting
discoveries such as the first million bp of a
Neandertal genome (Green, 2006). Another
important field of 454 is metagenomics, for example
uncovering the potential cause of the disappearance
of the honeybee (Cox-Foster, 2007).
Fig. 1. Maximum read length HTS platform.
ISME 2016 - Information Science and Management Engineering IV
320
ISME 2016 - International Conference on Information System and Management Engineering
320
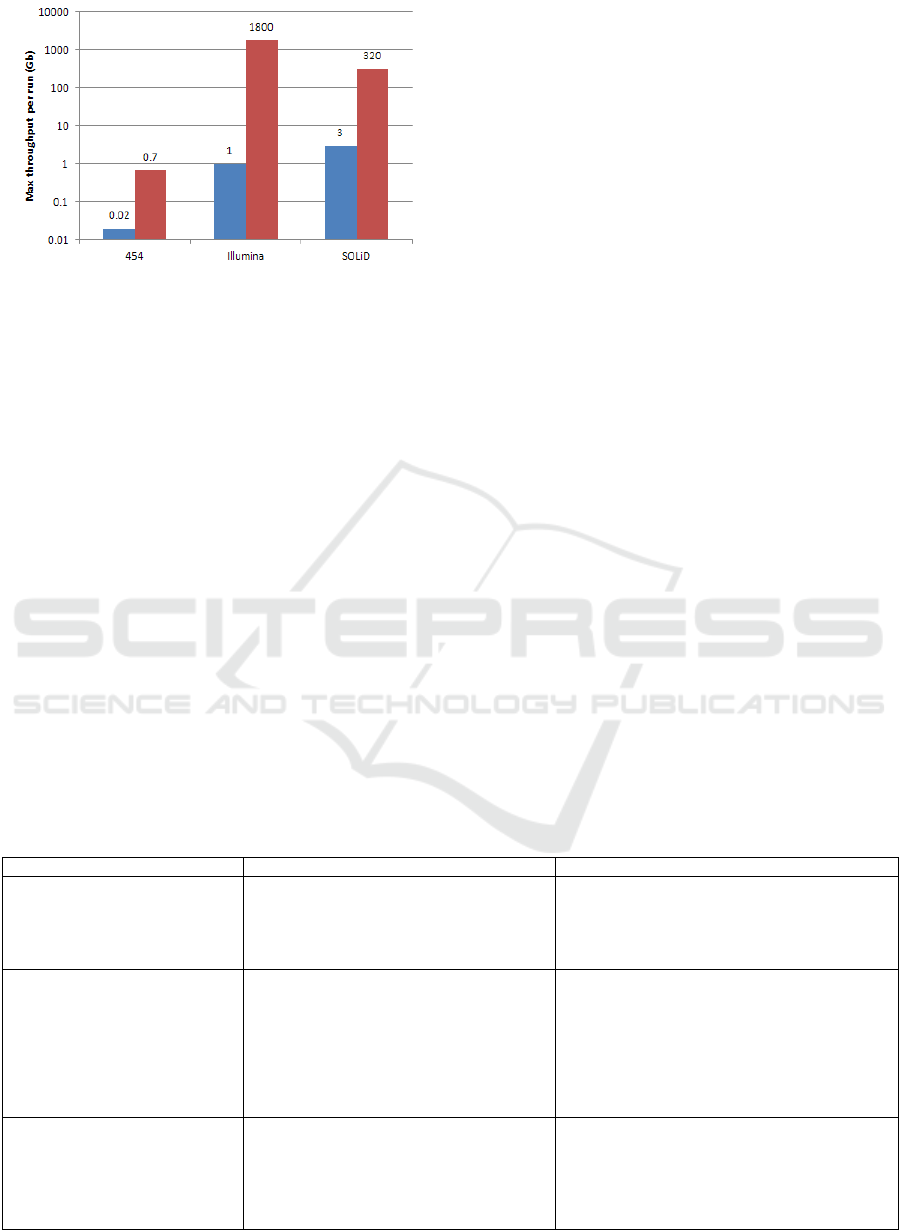
Fig. 2. Maximum throughput HTS platform.
With the rapid improvement of sequencing
machines and chemistries, Illumina technology can
now generate reads of several hundreds of bp long,
shown by Fig. 2. Thus, although reads produced by
Illumina still shorter than that produced by 454, de
novo genome assembly and metagenomics can now
also be performed with Illumina sequencing. Illumina
has been achieved remarkable increase in throughput,
which currently offers the highest throughput per run
with the lowest per base cost (Liu, 2012), shown by
Fig. 3. A summary of the advantages and drawbacks
of the different HTS systems is presented in Table 1.
3 THE APPLICATION OF HTS IN
HUMAN DISEASE RESEARCH
HTS is a new molecular detection technology with
high speed, high accuracy and high throughput. Its
research and application become more and more
widely in the field of life sciences and medical.
Genome data obtained by analysis and mining, and
genetic variation of information extracted from the
disease can provide a scientific basis for disease
diagnosis, treatment and prevention (Zaghloul,
2010).
In this part, the application of new generation
HTS in human disease is reviewed with exome
sequencing and Chip sequencing technology as an
example.
A.
Application of Exome Sequencing
in Human Diseases
Exome sequencing is a high-throughput sequencing
method by using special means to enrich the whole
exome. The basic processes include enrichment of
exome region sequences, high throughput
sequencing and bioinformatics analysis of
sequencing data.
The exon region contains the information
required by the synthesis of proteins, and covers
most of the functional variants associated with the
phenotype of the individual. Howerver, these exon
regions of the encoded protein account for only
about 1% of the human genome, thus it can greatly
improve the efficiency of exon region research
compared to the conventional PCR methods. In the
case of relatively high cost of sequencing, we can
obtain data with deeper coverage and higher
accuracy of sequencing coverage, and more coding
region information of the individual using the exome
sequencing under the same cost.
TABLE I. Pros and cons of the different HTS
Technology Pros Cons
Roche/454
The long reads (1 kb maximum) are easier to
map to a reference genome, and are an
advantage for de novo genome assemblies or
for Metagenomics applications. Run times
are relatively fast (~23 h)
Relatively low throughput (about 1 million reads,
700 Mb sequence data) and high reagent cost.
High error rates in homopolymer repeats .
Illumine/Solexa
Illumina is currently the leader in the HTS
industry and most library preparation
protocols are compatible with the Illumina
system. In addition, Illumina offers the
highest throughput of all platforms and the
lowest per-base cost. Read lengths of up to
300 bp, compatible with almost all types of
application.
Sample loading is technically challenging; owing
to the random scattering of clusters across the
flow cells library concentration must be tightly
controlled. Overloading results in overlapping
clusters and poor sequence quality.
SOLID
Second highest throughput system on the
market. The SOLiD system is widely
claimed to have lower error rates, 99.94%
accuracy, than most other systems owing to
the fact that each base is read twice.
Shortest reads (75 nt maximum) of all platforms,
and relatively long run times. Less-well-suited for
de novo genome assembly. The SOLiD system is
much less widely used than the Illumina system
and the panel of sample preparation kits and
services is less well developed.
High-Throughput Sequencing Technology and Its Applications in Human Disease
321
High-Throughput Sequencing Technology and Its Applications in Human Disease
321

Exome sequencing is an effective method to
detect pathogenicity genes and susceptibility loci at
the genomic level. It can not only rapidly locate the
gene of single gene disease, but also be used to study
the common diseases caused by mutations, such as
diabetes, hypertension and tumor.
Nat Genet et al. (Ng, 2009) reported on the
targeted capture and massively parallel sequencing
of the exomes of 12 humans. Using FSS as a proof-
of-concept, they showed that candidate genes for
Mendelian disorders could be identified by exome
sequencing of a small number of unrelated, affected
individuals. In the same year, Nat Genet et al. (Ng,
2010) demonstrated the first successful application
of exome sequencing to discover the gene for a rare
mendelian disorder of unknown cause. Jones et al.
(Jones, 2010) determined the exomic sequences of
eight tumors after immunoaffinity purification of
cancer cells. Through comparative analyses of
normal cells from the same patients, they identified
four genes that were mutated in at least two tumors,
PIK3CA, KRAS, PPP2R1A and ARID1A. The
nature and pattern of the mutations suggested that
PPP2R1A functions as an oncogene and ARID1A as
a tumor-suppressor gene. Harbour et al. (Harbour,
2010) used exome capture coupled with massively
parallel sequencing to search for metastasis-related
mutations in highly metastatic uveal melanomas of
the eye. Their findings implicated loss of BAP1 in
uveal melanoma metastasis and suggested that the
BAP1 pathway might be a valuable therapeutic
target. Seshagiri et al. (Seshagiri, 2012) analysed
systematically more than 70 pairs of primary human
colon tumours by applying next-generation
sequencing to characterize their exomes,
transcriptomes and copy-number alterations. Their
analysis for significantly mutated cancer genes
identified 23 candidates, including the cell cycle
checkpoint kinase ATM. Li et al. (Li, 2011)
discovered novel inactivating mutations of ARID2 in
four major subtypes of HCC. Notably, 18.2% of
individuals with HCV-associated HCC in the United
States and Europe harbored ARID2 inactivation
mutations, suggesting that ARID2 was a tumor
suppressor gene that was relatively commonly
mutated in this tumor subtype. Xu et al. (Xu, 2011)
examined the possibility that rare de novo protein-
altering mutations contribute to the genetic
component of schizophrenia by sequencing the
exomes of 53 sporadic cases, 22 unaffected controls
and their parents. They analyses suggested a major
role for de novo mutations in schizophrenia as well
as a large mutational target, which together provide a
plausible explanation for the high global incidence
and persistence of the disease. Glessner et al.
(Glessner, 2014) studied 538 CHD trios using
genome-wide dense single nucleotide polymorphism
arrays and whole exome sequencing. Integrating de
novo variants in whole exome sequencing and CNV
data suggested that ETS1 was the pathogenic gene
altered by 11q24.2-q25 deletions in Jacobsen
syndrome and that CTBP2 was the pathogenic gene
in 10q subtelomeric deletions.
B.
ChIP and its Application in Human
Disease
ChIP uses an immune reagent specific for a DNA
binding factor to enrich target DNA sites to which
the factor is bound in the living cell. The enriched
DNA sites are then identified and quantified.
Because of the gigabase-size genomes of vertebrates,
ChIP cannot combine high accuracy, whole-genome
completeness, and high binding-site resolution.
Chromatin immunoprecipitation followed by
sequencing, short for ChIPSeq, is a high-throughput
method combining ChIP with a HTS (Johnson,
2007). The ChIPSeq differs from other large-scale
ChIP methods such as ChIPchip in design, data
produced, and cost. ChIP-seq has the advantage of
high-resolution, low-noise, high coverage to study
the protein-DNA interactions (Schones, 2008).
ChIP-seq can be applied to any species with known
genome sequence, and study the interaction between
any kind of DNA-related protein and its Target
DNA.
ChIPSeq illustrates the power of new sequencing
platforms, such as those from Solexa/ Illumina and
454, to perform sequence census counting assays.
The generic task in these applications is to identify
and quantify the molecular contents of a nucleic acid
sample whose genome of origin has been sequenced
(Johnson, 2007).
With the reduction of sequencing costs, ChIP-seq
gradually becomes a common method of studying
gene regulation and epigenetic mechanism. The
applications of ChIP-seq in human disease research
are becoming more and more extensive.
Robertson et al. (Robertson, 2007) used ChIP-seq
to map STAT1 targets in interferon-bold gamma
(IFN-bold gamma)-stimulated and unstimulated
human HeLa S3 cells, and compared the method’s
performance to ChIP-PCR and to ChIP-chip for four
chromosomes. By ChIP-seq, using 15.1 and 12.9
million uniquely mapped sequence reads, and an
estimated false discovery rate of less than 0.001,
they identified 41,582 and 11,004 putative STAT1-
binding regions in stimulated and unstimulated cells,
ISME 2016 - Information Science and Management Engineering IV
322
ISME 2016 - International Conference on Information System and Management Engineering
322

respectively. Lin et al. (Lin, 2009) applied
expression profiling to identify the response program
of PC3 cells expressing the AR (PC3-AR) under
different growth conditions (i.e. with or without
androgens and at different concentration of
androgens) and then applied the newly developed
ChIP-seq technology to identify the AR binding
regions in the PC3 cancer genome. They found that
the comparison of MOCK-transfected PC3 cells with
AR-transfected cells identified 3,452 differentially
expressed genes (two fold cutoff) even without the
addition of androgens (i.e. in ethanol control),
suggesting that a ligand independent activation or
extremely low-level androgen activation of the AR.
ChIP-Seq analysis revealed 6,629 AR binding
regions in the cancer genome of PC3 cells with an
FDR (false discovery rate) cut off of 0.05. Hurtado
et al. (Hurtado, 2011) used ChIP-seq to research
breast cancers and found that FOXA1 was a key
determinant that can influence differential
interactions between estrogen receptor-α (ER) and
chromatin. Ross-Innes et al. (Ross-Innes, 2012)
mapped genome-wide ER-binding events, by
chromatin immunoprecipitation followed by ChIP-
seq, in primary breast cancers from patients with
different clinical outcomes and in distant ER-
positive metastases. Results showed that there was
plasticity in ER-binding capacity, with distinct
combinations of cis-regulatory elements linked with
the different clinical outcomes. Wang et al. (Wang,
2012) analyzed genome-wide occupancy patterns of
CTCF by ChIP-seq in 19 diverse human cell types,
including normal primary cells and immortal lines.
Results revealed a tight linkage between DNA
methylation and the global occupancy patterns of a
major sequence-specific regulatory factor. Lee et al.
(Lee, 2012) introduced genome-wide studies that
mappped the binding sites of CTCF and its
interacting partner, cohesin, using chromatin
immunoprecipitation coupled with ChIP-seq
revealded that CTCF globally co-localizes with
cohesin.
4 CONCLUSIONS
Ongoing cost reduction and the development of
standardized pipelines will probably make HTS a
standard tool for a multifaceted approach involving
clinical and research laboratories, bioinformatics
scientists, biotechnology companies, and regulatory
agencies in the near future.
Nevertheless, the implementation of HTS faces
significant challenges, in particular high data storage
and complex processing. With time goes on, the
amount of human genomes will be far more than the
already impressive amount of sequence data
available. Due to so many people’s genomes
sequenced, it is in great need to increase the data
storage capacity, speed up the establishment and
maintenance of databases, and develop efficient data
retrieval methods. For complex diseases, the
relationship between the massive sequencing
genome data and disease is not clear. Disease
pathogenic and development process cannot be
guaranteed only by obtained genomic information.
Biochemical data, such as transcriptome, proteome,
macro-genome, etc. as well as CT and MRI images
are needed to combine together to construct a large-
scale multi-dimensional life health data collection.
With the rapid development of cloud computing
technology, the storage, analysis and monitoring of
various vital signs and data will gradually be
realized. Accurate analysis and data mining will
decipher the causes of human disease and promote
the development of precision medicine.
REFERENCES
Chung W. J., Long J., Cheng J., et al., 2015, Next
generation sequencing analysis of genetically
engineered mouse models of human cancers, Cancer
Research, 75(15 Supplement): 2987-2987.
Comas I., Gil A., 2016, Next generation sequencing for the
diagnostics and epidemiology of tuberculosis,
Enfermedades Infecciosas y Microbiología Clínica,
34: 32.
Cox-Foster, D. L. et al., 2007, A metagenomic survey of
microbes in honey bee colony collapse disorder,
Science 318, 283-287.
Deeb K. K., Hohman C. M., Risch N. F., et al., 2015,
Routine clinical mutation profiling of non-small cell
lung cancer using next-generation sequencing,
Archives of Pathology and Laboratory Medicine,
139(7): 913-921.
Georgiou G., Ippolito G. C., Beausang J., et al., 2014, The
promise and challenge of high-throughput sequencing
of the antibody repertoire, Nature biotechnology,
32(2): 158-168.
Glessner J. T., Bick A. G., Ito K., et al., 2014, Increased
frequency of de novo copy number variants in
congenital heart disease by integrative analysis of
single nucleotide polymorphism array and exome
sequence data, Circulation research, 115(10): 884-
896.
Green, R. E. et al., 2006, Analysis of one million base
pairs of Neanderthal DNA, Nature 444, 330-336.
Gu Y., Lu K., Yang G., et al., 2014, Mutation spectrum of
six genes in Chinese phenylketonuria patients obtained
High-Throughput Sequencing Technology and Its Applications in Human Disease
323
High-Throughput Sequencing Technology and Its Applications in Human Disease
323

through next-generation sequencing, PLoS One, 9(4):
e94100- e94100.
Harbour J. W., Onken M. D., Roberson E. D. O., et al.,
2010, Frequent mutation of BAP1 in metastasizing
uveal melanomas, Science, 330(6009): 1410-1413.
Hurtado A., Holmes K. A., Ross-Innes C. S., et al., 2011,
FOXA1 is a key determinant of estrogen receptor
function and endocrine response, Nature genetics,
43(1): 27-33.
Johnson D. S., Mortazavi A., Myers R. M., et al., 2007,
Genome-wide mapping of in vivo protein-DNA
interactions, Science, 316 (5830): 1497-1502.
Jones S., Wang T. L., Shih I. M., et al., 2010, Frequent
mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma, Science, 330(6001): 228-
231.
Krauskopf J., Caiment F., Claessen S. M., et al., 2015,
Application of high-throughput sequencing to
circulating microRNAs reveals novel biomarkers for
drug-induced liver injury, Toxicological Sciences,
143(2): 268-276.
Lee B. K., Iyer V. R., 2012, Genome-wide studies of
CCCTC-binding factor (CTCF) and cohesin provide
insight into chromatin structure and regulation,
Journal of Biological Chemistry, 287(37): 30906-
30913.
Li M., Zhao H., Zhang X., et al., 2011, Inactivating
mutations of the chromatin remodeling gene ARID2 in
hepatocellular carcinoma, Nature genetics, 43(9): 828-
829.
Lin B., Wang J., Hong X., et al., 2009, Integrated
expression profiling and ChIP-seq analyses of the
growth inhibition response program of the androgen
receptor, PloS one, 4(8): e6589.
Liu, L. et al., 2012, Comparison of next-generation
sequencing systems, J. Biomed. Biotechnol, 251364.
Mardis E. R., 2008, Next-generation DNA sequencing
methods. Annu Rev Genomics Hum Genet, 9: 387-402.
Ng S. B., Turner E. H., Robertson P. D., et al., 2009,
Targeted capture and massively parallel sequencing of
12 human exomes, Nature, 461(7261): 272-276.
Ng S. B., Buckingham K. J., Lee C., et al., 2010, Exome
sequencing identifies the cause of a mendelian
disorder, Nature genetics, 42(1): 30-35.
Norton M. E., Jacobsson B., Swamy G. K., et al., 2015,
Cell-free DNA analysis for noninvasive examination
of trisomy, New England Journal of Medicine,
372(17): 1589-1597.
Park, P. J., 2009, ChIP-seq: advantages and challenges of a
maturing technology, Nat. Rev. Genet, 10: 669-680.
Redin C., Gérard B., Lauer J., et al., 2014, Efficient
strategy for the molecular diagnosis of intellectual
disability using targeted high-throughput sequencing.
Journal of medical genetics, jmedgenet-2014-102554.
Renkema K. Y., Stokman M. F., Giles R. H., et al., 2014,
Next-generation sequencing for research and
diagnostics in kidney disease, Nature Reviews
Nephrology, 10(8): 433-444.
Reuter J. A., Spacek D. V., Snyder M. P., 2015. High-
throughput sequencing technologies, Molecular cell,
58(4): 586-597.
Robertson G., Hirst M., Bainbridge M., et al., 2007,
Genome-wide profiles of STAT1 DNA association
using chromatin immunoprecipitation and massively
parallel sequencing, Nature methods, 4(8): 651-657.
Ross-Innes C. S., Stark R., Teschendorff A. E., et al., 2012,
Differential oestrogen receptor binding is associated
with clinical outcome in breast cancer, Nature, 481
(7381): 389-393.
Saare M., Rekker K., Laisk-Podar T., et al., 2014, High-
throughput sequencing approach uncovers the
miRNome of peritoneal endometriotic lesions and
adjacent healthy tissues, PloS one, 9(11): e112630.
Schones D. E., Zhao K., 2008, Genome-wide approaches
to studying chromatin modifications, Nat Rev Genet,
9(3): 179-191.
Seshagiri S., Stawiski E. W., Durinck S., et al., 2012,
Recurrent R-spondin fusions in colon cancer, Nature,
488(7413): 660-664.
Tawfik D. S., Griffiths A. D., 1998, Man-made cell-like
compartments for molecular evolution, Nat
Biotechnol, 16 (7): 652-656.
Thibodeau B., Cardenas P. Y., Ahmed S., et al., 2016,
Next generation sequencing of brain metastasis in non-
small cell lung cancer, Cancer Research, 76(14
Supplement): 1535-1535.
Van Dijk E. L., Auger H., Jaszczyszyn Y., et al., 2014,
Ten years of next-generation sequencing technology,
Trends in genetics, 30(9): 418-426.
Wang H., Maurano M. T., Qu H., et al., 2012, Widespread
plasticity in CTCF occupancy linked to DNA
methylation, Genome research, 22(9): 1680-1688.
Wang, Z., et al., 2009, RNA-Seq: a revolutionary tool for
transcriptomics, Nat. Rev. Genet, 10: 57-63.
Wilson K. D., Shen P., Fung E., et al., 2015, A rapid, high-
quality, cost-effective, comprehensive, and expandable
targeted next-generation sequencing assay for
inherited heart diseases, Circ Res, 117(7): 603-611.
Wolf Z. T., Brand H. A., Shaffer J. R., et al., 2015,
Genome-wide association studies in dogs and humans
identify ADAMTS20 as a risk variant for cleft lip and
palate, PLoS Genet, 11(3): e1005059-e1005059.
Xu B., Roos J. L., Dexheimer P., et al., 2011 Exome
sequencing supports a de novo mutational paradigm
for schizophrenia, Nature genetics, 43(9): 864-868.
Xu Y., Chen S., Yin S., et al., 2015, Embryo genome
profiling by single-cell sequencing for preimplantation
genetic diagnosis in a beta-thalassemia family, Clin
Chem
, 61(4): 617-626.
Zaghloul N. A., 2010, Katsanis N. Functional modules,
mutational load and human genetic disease, Trends
Genet, 26(4): 168-76.
ISME 2016 - Information Science and Management Engineering IV
324
ISME 2016 - International Conference on Information System and Management Engineering
324
