
Ensemble Learning-based Prediction of Drug-pathway Interactions
based on Features Integration
Mingyuan Xin
1
, Jun Fan
2
and Zhenran Jiang
2
1
Shanghai Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences and School of Life Sciences,
East China Normal University, Shanghai 200241, China
2
Shanghai Key Laboratory of Multidimensional Information Processing, Department of Computer Science and Technology,
East China Normal University, Shanghai 200241, China
Keywords: Drug-pathway Interaction, Ensemble Learning, AdaBoost, Bagging, Random SubSpace.
Abstract: Recently, developing computational methods to explore drug-pathway interaction relationships has attracted
attention for their potentiality in discovering unknown targets and mechanisms of drug actions. However,
mining suitable features of drugs and pathways is challenging for available prediction methods. This paper
performed an ensemble learning-based method to predict potential drug-pathway interactions by integrating
different drug-based and pathway-based features. The main characteristic of our method lies in using the
Relief algorithm for feature selection and regarding three ensemble methods (AdaBoost, Bagging and
Random Subspace) for classifiers. Cross validation results showed the AdaBoost algorithm that based on the
Decision Tree classifier can obtain a higher prediction accuracy, which indicated the effectiveness of
ensemble learning. Moreover, some new predicted interactions were validated by database searching, which
demonstrated its potentiality for further biological experiment investigation.
1 INTRODUCTION
Traditional drug discovery primarily tries to seek the
specific drug molecule to act on individual target
(Hopkins, 2008). However, it is well recognized that
many drugs are far beyond targeting individual
proteins, but rather influencing the complex
interactions among the relevant biological pathways.
Therefore, the inferences of drug-pathway
associations are critical for identifying unknown
targeted pathways and drug action mechanisms (Ma
and Zhao, 2012).
Increasing effort has been devoted to detecting
these potential associations and several drug-
pathway interactions prediction methods have been
proposed from different aspects (Subramanian et al.,
2005; Ma and Zhao, 2012). Generally, most of the
methods attempted to analyze the drug-pathway
interactions mainly based on gene expression data.
For instance, ‘iFad’ mainly combined the gene
expression and drug sensitivity datasets to analyze
the drug-pathway interactions (Ma and Zhao, 2012),
but it is always difficult to obtain adequate drug-
pathway information merely on the gene expression
data. To tackle the problem, some methods attempt
to utilize different machine learning algorithms by
integrating more chemical and biological
information (Silberberg et al., 2012; Pratanwanich
and Lio, 2014; Song et al., 2014). For instance,
protein-protein interaction networks (PPI)
(Silberberg et al., 2012), other target structure
information have been utilized effectively recently.
However, the extraction and fusion of the drug-
pathway association information is still challenging
for drug-pathway interactions prediction (Song et
al., 2014).
Inspired by the challenges, we attempted to use
the ensemble learning methods to predict potential
drug-pathway associations. As similar drugs often
act similar target proteins, we assume that similar
drugs also act on similar pathways. Based on the fact
that the drug mode of actions (MoA) is a central
concept linking drug structures to a set of biological
activities, we used drug structure and MoA
similarity to represent drug feature information.
Further, we used the ‘RNA: AffyHG-U133 (A, B)’
gene expression data of NCI-60 cell lines (Reinhold
et al., 2012) to obtain related genes which covered
by these pathways, then these genes ontology
semantic similarity and sequence similarity are
Xin M., Fan J. and Jiang Z.
Ensemble Learning-based Prediction of Drug-pathway Interactions based on Features Integration.
DOI: 10.5220/0006096701170124
In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2017), pages 117-124
ISBN: 978-989-758-214-1
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
117

calculated to represent pathway information.
Further, the drug-pathway network topology
information was merged into the drug and pathway
feature profiles, respectively.
It is known that ensemble learning methods
usually exhibits better generalization performance
than a single classifier. In this study, we used three
well-established methods: AdaBoost (Freund and
Schapire, 1997), Bagging (Breiman, 1996) and
Random SubSpace (Ho, 1998) to achieve a good
ensemble result. Meanwhile, three widely used
learning methods: Support Vector Machine (SVM)
(Cortes and Vapnik, 1995), Navie Bayesian (NB)
(Rish, 2001) and Decision Tree (DT) (Friedl and
Brodley, 1997) are chosen as the base classifier.
Compared of these method combinations, the
AdaBoost algorithm that based on the DT classifier
is selected as the final model to predict the drug-
pathway interactions.
2 MATERIAL AND METHOD
2.1 Dataset
This study focus on 58 pathways that have been
proved to be related to cancers (Ahmed et al., 2011)
and 362 drugs obtained from KEGG database
(Kanehisa et al., 2012), which contains most of
pathways and molecular information in genomics,
transcriptomics, proteomics and metabolomics. In
addition, these drugs have complete drug
information and most of them are proved to be
related to these pathways.
2.1.1 Features Construction
(1) Drug features
Drug structure-based feature
d
S : Drug structure
similarity is calculated based on their molecular
fingerprints which include 881 chemical
substructures defined by the PubChem database
(Chen et al., 2009). PaDel-Descriptor (Yap, 2011)
was used to convert each drug Mol file into 881
dimensional binary vectors. Then the corresponding
fingerprints are used to compute the similarity scores
between two drugs by Tanimoto scores (Lipkus,
1999).
MoA based feature
d
F
: Since drugs which share a
similar MoA are likely to target same pathways, thus
the drug MoA similarity can be utilized to predict
associations between drugs and pathways. Here we
retrieved MoA information from DrugBank database
(Wishart et al., 2006) and calculate the similarities
based on 341 MoAs. We consider drugs as samples
and each MoA as a label and take known drug-MoA
association matrix M as local correlations.
According to the local correlations between labels of
samples in drug-MoA interaction network, we
calculate the cosine similarity of each two drug
vectors in M:
(, ) cos( , )
|| |||| ||
T
ij
dij
ij
mm
Sij mm
mm
.
(1)
(2)Pathway features
This study mainly concentrated on 1863 genes
covered by the 58 pathways for the pathway features
construction.
Gene ontology Semantic feature
p
F
: The Gene
Ontology terms of 1863 genes were retrieved from
Quick GO database (Binns et al., 2009), and
semantic similarity scores between these pathway-
related genes were calculated by the csbl.go R
package (Ovaska et al., 2008). What’s more, the
similarity scores between the pathways from gene
semantic similarity scores were computed in
accordance with the reference (Song et al., 2014).
Gene sequence similarity
p
S
: Sequence similarity
between the corresponding pathway-related genes
was calculated based on a Smith-Waterman
sequence alignment score (Smith et al., 1985), and
the similarity between two pathways can be
calculated as the sum of similarity between all the
gene sequences related to the two pathways.
(3)Drug-pathway network topology feature
The drug-pathway network topology information
was calculated based on the literature (Van et al.,
2013). In the drug-pathway network, the average
shortest path of each node and the number of shared
drugs or pathways are denoted as
,,,
dpdp
D
DKK
,
respectively.
As showed in Table 1, the drug features
d
Sim
include drug structure information
d
S , drug mode of
actions
d
M
, network topology information
,
dd
DK
,
and the pathway features
p
Sim
are combined by
gene ontology semantic similarity
p
G
, gene
sequence similarity
p
S
and ,
p
p
DK. Construction of
the drug-pathway feature is followed the theory: for
drug i and pathway j, their features can be
constructed by combining row i in
d
Sim and row j
in
p
Sim
, namely.
Fea<drug(i),pathway(j)>=
d
Sim (i)+
p
Sim
(j). (2)
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
118

Table 1: The construction of drug-pathway features.
Drug-Pathway Features
Drug
Features
d
Sim
Drug structure similarity
d
S
Drug mode of actions
similarity
d
M
Drug-pathway interaction
topology information
,
dd
DK
Pathway
Features
p
Sim
Pathway-related gene
ontology semantic feature
p
G
Pathway-related gene
sequence similarity
p
S
Drug-pathway interaction
topology information
,
p
p
DK
2.1.2 Features Selection
Existing facts demonstrate that irrelevant and
redundant features can lead the model to overfit.
Here we perform the Relief method (Sun et al.,
2011) to avoid redundancy of feature variables. At
each iteration, the algorithm picks randomly a
sample K, then picks at random the feature sample
of the instance closest to K from each class, the
same class instance is called ‘near-hit’ and the
different class instance is called ‘near-miss’. Then
the weight vector is updated as:
22
()( )
iii i i i
W W x nearHit x nearMiss
(3)
Thus, the weight of any given feature increases if
the distance between K and near-hit is shorter than
the distance between K and near-miss for the feature,
and decreases otherwise. After n iterations, the
relevance vector is updated by dividing each element
of the weight vector by n, then feature are selected if
their relevance is greater than a threshold k. In this
study, we set the threshold as zero and finally
selected 551 features with positive weight from 764
features.
2.2 Ensemble Learning Method
Ensemble learning is a machine learning paradigm
which constructs a set of classifiers and then
combines them for classifying data by taking a vote
of their predictions (Schwenker, 2013). Here we take
three well-established methods in practice to achieve
a good ensemble. AdaBoost and Bagging are two
instance partitioning methods and Random Subspace
is a feature partitioning method (Van et al., 2013).
2.2.1 AdaBoost
AdaBoost is an iterative algorithm where the
conjuncture of many weak classifiers is employed to
construct a ‘strong’ classifier (Ho, 1998). It works
by choosing a base algorithm and iteratively
improving it by accounting for the incorrectly
classified examples in the training set. The final
predictions are retrieved from a weighted vote. The
AdaBoost algorithm’s pseudo code is shown as
followed:
2.2.2 Bagging
Bagging is an ensemble meta-estimator where each
base classifier is trained on random subsets of the
original dataset and then aggregated their individual
predictions to form a final prediction (Breiman,
1996). It improves the stability and reduces variance,
and avoids overfitting of learning algorithms. The
base classifiers’ combination strategy for Bagging is
majority vote. The Bagging algorithm pseudo code
is shown as followed:
2.2.3 Random Subspace
Random Subspace is a combination model that
consists of several classifiers and each are trained on
randomly chosen subspaces of the original feature
space (Ho, 1998). The outputs of the models are
usually combined by majority vote. The Random
Subspace algorithm’s pseudo codes are shown as
followed:
Ensemble Learning-based Prediction of Drug-pathway Interactions based on Features Integration
119

2.3 Procedure
In this model, we choose four widely used base
classifier for implementing the three ensemble
methods: SVM, NB and
DT. SVM algorithm has
been used for a variety of application and it performs
structural risk minimization on a nested set structure
of separating hyperplanes (Cortes and Vapnik, 1995).
Navie Bayesian algorithm is a simple classification
based on the Bayes theory for conditional
probability. Decision Tree algorithm is an easily
understandable and transparent sequential model but
it has relatively low prediction accuracy compared to
other methods. In this study, we chose the widely
used method C4.5. Here we use the toolkit WEKA,
which includes a collection of machine learning
algorithms for solving data mining problems (Hall et
al., 2009). The AdaBoost, Bagging and Random
SubSpace are selected to implement the ensemble
algorithms. The drug-pathway associations we used
include 643 positive samples and 17390 negative
samples, and the positive sample density of the
dataset is 0.036.
In order to evaluate the performances of different
models, 10-fold cross validation tests are executed
on the models. For the datasets, all of the drug-
pathway samples are randomly spilt into ten subsets
with equal size, and nine subsets are combined as
the training set and the remaining one subset is taken
as the testing set each time. The overview procedure
of the model is shown in the Fig.1.
Figure 1: Figure summarizes the overview of this model.
The model is mainly composed of three sections: (a) the
process of feature construction. (b) ensemble methods
operation. (c) the comparison of these methods.
3 RESULTS
3.1 Performance Evaluation
Here several metrics, i.e., precision, recall, accuracy
(ACC), area under ROC curve (AUC) and the area
under the precision-recall curve (AUPR), F-measure
(F), are used to evaluate the performances of the
models. Among the metrics, accuracy represents the
overall accuracy of the classification, precision
represents the measure of the reliability of positive
instances prediction and recall represents the
probability of correct prediction. F-measure is a
score from 0 to 1 as a measure of test accuracy. The
metrics were calculated in a 10-fold cross-validation
procedure by using the equations as followed:
()
2
TP
precision
TP FP
TP
recall
TP FN
TP TN
ACC
TP TN FP FN
p
recision recall
F
p
recision recall
(4)
where TP, FP, TN and FN represent the number
of true positive, false positive, true negative and
false negative samples, respectively.
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
120

3.2 Performance of Features
Integration
To quantitatively assess the efficiency of all the
features and each single feature in predicting the
drug-pathway interactions, we performed a 10-fold
cross validation with the AdaBoost algorithm based
on DT classifier, respectively. As a result, the model
that integrated features exhibits a better performance
than those with single feature (See in Fig. 2).
Further, the Relief method is performed to avoid
the redundancy of feature variables. In our study, we
get 551 features with positive weight from 764
features after feature selection. By comparison, the
selected features have better classification
performances than the original features (See in
Fig.3).
3.3 Performance of Ensemble Methods
In this model, we compared the performance of 12
methods, including SVM, NB and DT, and their
corresponding ensemble methods of AdaBoost,
Bagging and Random Subspace. The performance of
base classifiers and ensemble methods based on
three base classifiers is shown in Table 2. As
demonstrated in Table 2, we find all the three base
classifiers have a poorer performance than the
ensemble methods, and AdaBoost method has the
best performance in every base classifier. The
possible reason for this situation is that AdaBoost
more fully account the weight of each classifier
relative to other algorithms.
Next, we compared the three base classifiers in
the case of AdaBoost ensemble models. The ROC
and PR curves of the three approaches are shown in
the Fig. 4, we can see that the AdaBoost ensemble
algorithm based on the DT classifier can achieve the
best performance.
3.4 New Predictions
Here we used the Comparative Toxicogenomics
Database (CTD) (Davis et al., 2015) as reference to
validate the predicted interactions. The CTD
database integrates chemical, gene, disease and their
interactions from curated literatures. There are 502
new predicted interactions and 241 associations have
been proved existence by searching the CTD
database. For instance, the interaction between the
drug ‘Theophylline’ and the pathway ‘Neuroactive
ligand-receptor interaction’ can be found in both
KEGG database and CTD database. Some predicted
samples that have been confirmed in CTD database
are listed in Table 3.
In addition, we focused on the pathway:
Kegg05223 and associated predicted drugs. We
found there are 15 predicted drugs related with the
pathway ‘Non-small cell lung cancer’. Meanwhile,
we confirmed that eleven drugs have associations
with the pathway in CTD database. Among the other
four drugs, we cannot find the interactions between
the drug and the pathway ‘Non-small cell lung
cancer’, but from the aspect of disease we find that
the three drug ‘Aminoglutethimide’, ‘Sunitinib
malate’ and ‘sunitinib’ have been tested in clinical
trials for lung cancer in the literatures (Xiao et al.,
2010; Chen et al., 2011; Shin et al., 2013; Xue et al.,
2014), which have been laterally validated that the
drugs have associations with this pathway.
Figure 2: The comparison between the integrated features and each single feature.
Ensemble Learning-based Prediction of Drug-pathway Interactions based on Features Integration
121
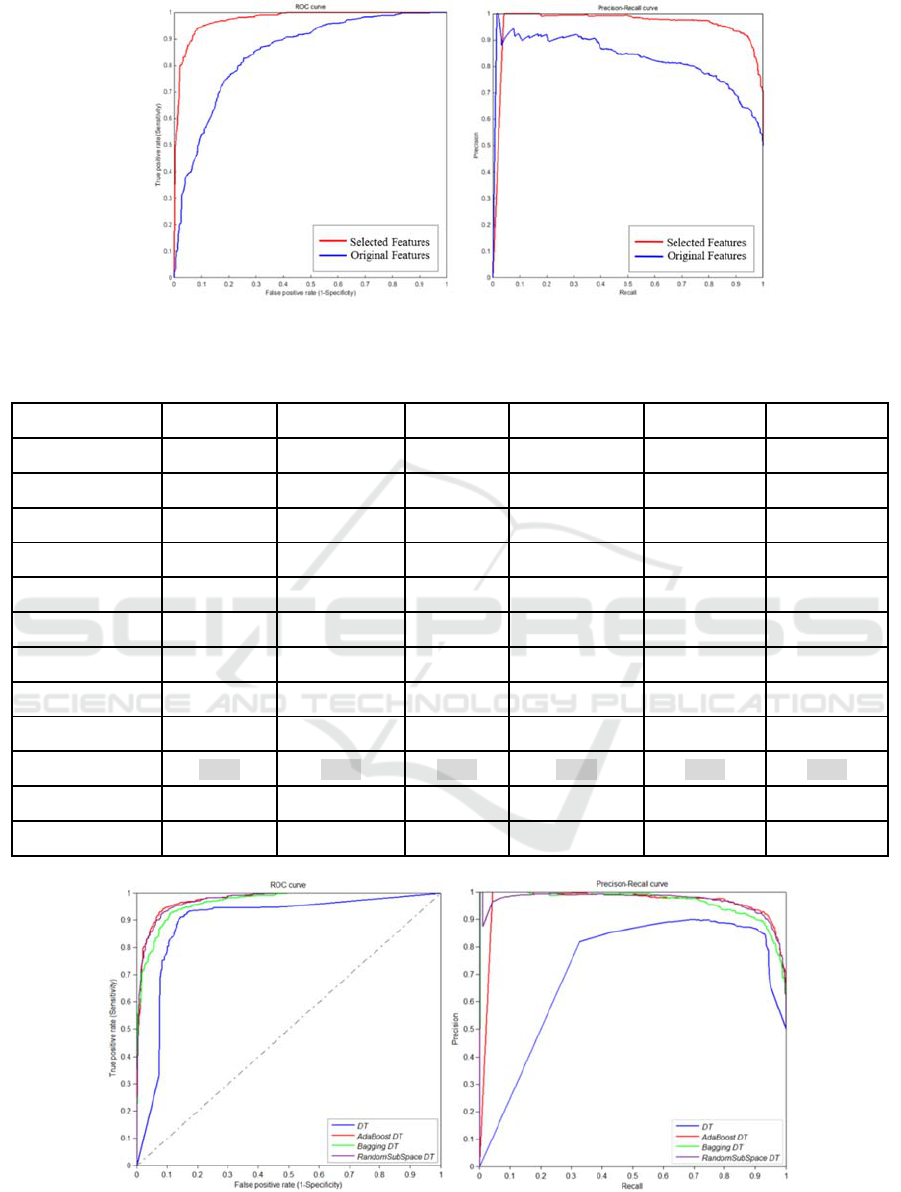
Figure 3: The comparison between selected features and original features.
Table 2: Performance comparisons of different learning methods.
Method AUC AUPR Recall Precision ACC F
SVM 0.827 0.770 0.827 0.827 0.653 0.827
AdaBoost SVM 0.922 0.925 0.834 0.835 0.669 0.834
Bagging SVM 0.856 0.811 0.826 0.826 0.652 0.826
RS SVM 0.859 0.821 0.804 0.804 0.608 0.804
NB 0.772 0.732 0.715 0.715 0.429 0.715
AdaBoost NB 0.851 0.845 0.779 0.779 0.558 0.778
Bagging NB 0.813 0.792 0.725 0.726 0.451 0.725
RS NB 0.804 0.790 0.708 0.709 0.418 0.708
DT 0.891 0.857 0.882 0.883 0.765 0.882
AdaBoost DT 0.975 0.976 0.925 0.925 0.850 0.925
Bagging DT 0.965 0.966 0.901 0.902 0.803 0.900
RS DT 0.974 0.972 0.916 0.916 0.833 0.916
Figure 4: The evaluation of the four methods: DT, AdaBoost DT, Bagging DT and Random SubSpace DT.
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
122
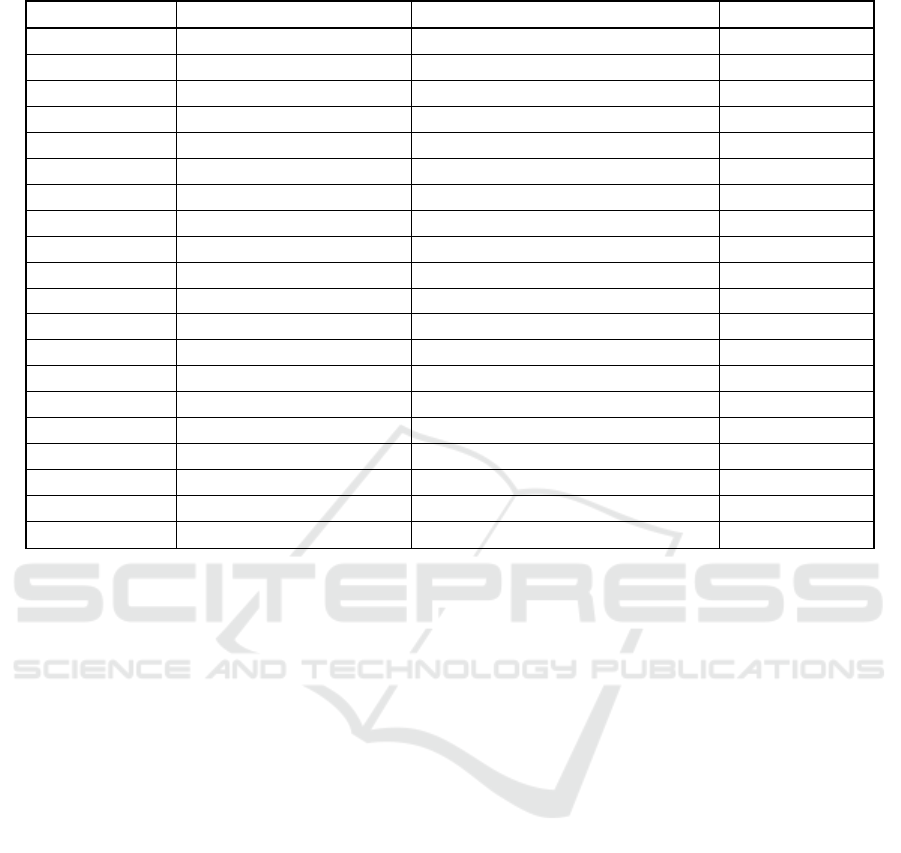
Table 3: The top 20 confirmed drug-pathway interactions.
DrugID (Kegg) Drug Name Pathway Name Validated Database
D00371 Theophylline Neuroactive ligand-receptor interaction Kegg; CTD
D04197 Floxuridine Natural killer cell mediated cytotoxicity CTD
D04023 Erlotinib hydrochloride Chronic myeloid leukemia CTD
D03881 Dobutamine tartrate Vascular smooth muscle contraction CTD
D03879 Dobutamine Vascular smooth muscle contraction CTD
D00371 Theophylline Vascular smooth muscle contraction Kegg; CTD
D00632 Dobutamine hydrochloride Vascular smooth muscle contraction CTD
D08111 Lercanidipine Vascular smooth muscle contraction Kegg; CTD
D01849 Lercanidipine hydrochloride Vascular smooth muscle contraction Kegg; CTD
D00126 Ibuprofen Insulin signaling pathway CTD
D01366 Bezafibrate Insulin signaling pathway CTD
D00341 Hydroxycarbamide Natural killer cell mediated cytotoxicity CTD
D00330 Flurbiprofen Insulin signaling pathway CTD
D00565 Fenofibrate Insulin signaling pathway CTD
D00586 Flutamide Non-small cell lung cancer CTD
D04023 Erlotinib hydrochloride Pancreatic cancer CTD
D00562 Propylthiouracil Natural killer cell mediated cytotoxicity CTD
D02368 Gemcitabine Cytokine-cytokine receptor interaction CTD
D01441 Imatinib mesilate Non-small cell lung cancer CTD
D01155 Gemcitabine hydrochloride Jak-STAT signaling pathway CTD
4 CONCLUSIONS
In this article, we evaluated the ensemble
algorithms: AdaBoost, Bagging and Random
SubSpace, for predicting drug-pathway interactions
based on three base classifiers: SVM, NB and DT.
Our results show that ensemble methods have the
advantage over the individual classifier on drug-
pathway interactions prediction. The merit of this
study lied in selecting the effective features obtained
from drug chemical structure information, drug
mode of actions and pathway-related gene
information. Some validated results to some extent
demonstrated the reliability of the models.
Although our method has utilized different types
of drug-based and pathway-based information, more
useful drug-pathway information can be further
mined. Therefore, our future study will focus on
fusing more biological prior information to improve
the prediction reliability.
ACKNOWLEDGEMENTS
This work was partly supported by National Natural
Science Foundation of China (Grants No.
81330049), National Major Scientific and
Technological Special Project for "Significant New
Drugs Development" (2013ZX09507001) and the
Science and Technology Commission of Shanghai
Municipality (14DZ2270100).
REFERENCES
Ahmed, J., Meinel, T., Dunkel, M., Murgueitio, M.S.,
Adams, R., Blasse, C., et al. 2011. CancerResource: a
comprehensive database of cancer-relevant proteins
and compound interactions supported by experimental
knowledge. Nucleic Acids Res., 39, D960-D967.
Binns, D., Dimmer, E., Huntley, R., Barrell, D.,
O'Donovan, C. and Apweiler, R., 2009. QuickGO: a
web-based tool for Gene Ontology searching.
Bioinformatics, 25(22), 3045-6.
Breiman, L., 1996. Bagging predictors. Machine Learning,
24(2), 123-140.
Chen, B., Wild, D. and Guha, R., 2009. PubChem as a
source of polypharmacology. J. Chem. Inf. Model., 49
(9), 2044-2055.
Chen, C., Fu, X., Zhang, D., Li, Y., Xie, Y., Li, Y. and
Huang Y., 2011. Varied pathways of stage IA lung
adenocarcinomas discovered by integrated gene
expression analysis. Int. J. Biol. Sci., 7(5), 551-66.
Ensemble Learning-based Prediction of Drug-pathway Interactions based on Features Integration
123

Cortes, C. and Vapnik, V., 1995. Support vector machine.
Machine Learning, 20(3), 273-297.
Davis, A.P., Grondin, C.J., Lennon-Hopkins, K., Saraceni-
Richards, C., Sciaky, D., King, B.L., et al. 2015. The
Comparative Toxicogenomics Database's 10th year
anniversary: update 2015. Nucleic Acids Res., 43,
D914-20.
Dietterich, T.G., 2000. Ensemble methods in machine
learning. In: Multiple classifier systems. Springer
Berlin Heidelberg, 1-15.
Freund, Y. and Schapire, R. E., 1997. A decision-theoretic
generalization of on-line learning and an application to
boosting. J Comput. Syst. Sci., 55(1), 119-139.
Friedl, M.A. and Brodley, C.E., 1997. Decision tree
classification of land cover from remotely sensed data.
Remote Sens. Environ., 61(3), 399-409.
Hall, M., Frank, E., Holmes, G., Pfahringer, B.,
Reutemann, P. and Witten, I. H., 2009. The WEKA
data mining software: an update. ACM SIGKDD
explorations newsletter, 11(1), 10-18.
Ho, T.K., 1998. The random subspace method for
constructing decision forests. IEEE Transactions on
Pattern Analysis and Machine Intelligence, 20(8),
832-844.
Hopkins, A.L., 2008. Network pharmacology: the next
paradigm in drug discovery. Nat Chem Biol, 4(11),
682-90.
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M. and
Tanabe, M., 2012. KEGG for integration and
interpretation of large-scale molecular data sets.
Nucleic Acids Res., 40, D109-D114.
Lipkus, A.H., 1999. A proof of the triangle inequality for
the Tanimoto distance. J Math Chem., 26, 263-265.
Ma, H. and Zhao, H., 2012. iFad: an integrative factor
analysis model for drug-pathway association
inference. Bioinformatics, 28(14), 1911-1918.
Ma, H. and Zhao, H., 2012. FacPad: Bayesian sparse
factor modeling for the inference of pathways
responsive to drug treatment. Bioinformatics, 28(20),
2662-70.
Ovaska, K., Laakso, M. and Hautaniemi, S., 2008. Fast
Gene Ontology based clustering for microarray
experiments. BioData Min., 1(1), 11.
Pratanwanich, N. and Lio, P., 2014. Exploring the
complexity of pathway–drug relationships using latent
Dirichlet allocation. Comput. Biol. Chem., 53,144-152.
Reinhold, W.C., Sunshine, M., Liu, H., Varma, S., Kohn,
K.W., Morris, J., et al. 2012. CellMiner: a web-based
suite of genomic and pharmacologic tools to explore
transcript and drug patterns in the NCI-60 cell lineset.
Cancer Res., 72(14), 3499-3511.
Rish, I., 2001. An empirical study of the naive Bayes
classifier. In: IJCAI 2001 workshop on empirical
methods in artificial intelligence, Hoos, H.H. Ed., IBM
New York, 3(22), pp. 41-46.
Schwenker, F., 2013. Ensemble methods: Foundations and
algorithms. Computational Intelligence Magazine,
IEEE, 8(1), 77-79.
Shin, J.Y., Hong, S.H., Kang, B., Minai-Tehrani, A. and
Cho, M.H., 2013. Overexpression of beclin1 induced
autophagy and apoptosis in lungs of K-rasLA1 mice.
Lung Cancer, 81(3), 362-70.
Silberberg, Y., Gottlieb, A., Kupiec, M., Ruppin, E. and
Sharan, R., 2012 Large-scale elucidation of drug
response pathways in humans. J. Comput. Biol., 19(2),
163-74.
Smith, T.F., Waterman, M.S. and Burks, C., 1985. The
statistical distribution of nucleic acid similarities.
Nucleic Acids Res., 13, 645-656.
Song, M., Yan, Y. and Jiang, Z., 2014. Drug-pathway
interaction prediction via multiple feature fusion. Mol.
Biosyst., 10(11), 2907-2913.
Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee,
S., Ebert, B.L., Gillette, M.A., et al. 2005. Gene set
enrichment analysis: a knowledge-based approach for
interpreting genome-wide expression profiles. Proc.
Natl. Acad. Sci. U. S. A., 102(43), 15545-50.
Sun, Y., Lou, X. and Bao, B., 2011. A novel relief feature
selection algorithm based on mean-variance model. J
Inf Comput Sci., 8, 3921-3929.
Van, L.T., Nabuurs, S.B. and Marchiori, E., 2013.
Predicting drug-target interaction networks of human
diseases based on multiple feature information.
Pharmacogenomics, 14(14), 1701-7.
Wishart, D.S., Knox, C., Guo, A.C., Shrivastava, S.,
Hassanali, M., Stothard, P., et al. 2006. DrugBank: a
comprehensive resource for in silico drug discovery
and exploration. Nucleic Acids Res., 34, D668-72.
Xiao, G., Lu, Q., Li, C., Wang, W., Chen, Y. and Xiao, Z.,
2010. Comparative proteome analysis of human
adenocarcinoma. Med Oncol., 27(2), 346-56.
Xue, D., Lu, M., Gao, B., Qiao, X. and Zhang, Y., 2014.
Screening for transcription factors and their regulatory
small molecules involved in regulating the functions
of CL1-5 cancer cells under the effects of
macrophage-conditioned medium. Oncol. Rep., 31(3),
23-33.
Yap, C.W., 2011. PaDEL-descriptor: An open source
software to calculate molecular descriptors and
fingerprints. J. Comput. Chem., 32(7), 1466-1474.
BIOINFORMATICS 2017 - 8th International Conference on Bioinformatics Models, Methods and Algorithms
124
