
A Network of Networks to Reproduce the Electrical Features of an
Aptamer-ligand Complex
What an Electrical Network Tells about Affinity
Eleonora Alfinito
1
, Rosella Cataldo
2
and Lino Reggiani
2
1
Dipartimento di Ingegneria dell’Innovazione, Salento University, Lecce, Italy
2
Dipartiemnto di Matematica e Fisica “Ennio de Giorgi”, Salento University, Lecce, Italy
Keywords: Networks, Proteotronics, Aptamers, Aptasensors.
Abstract: The increasing interest in the production and selection of aptamers for therapeutic and diagnostic applications
yields many studies in recent years. Most of them investigated the production techniques, usually performed
in vitro, but also the possibility of an in silico selection. Due to their specific ability of target-inhibition, some
aptamers are under clinical trials, and some other were just patented by several pharmaceutical companies.
However, the mechanism of aptamer-ligand formation is not completely understood. In this paper we explore
the possibility to describe some topological and electrical features of the aptamer TBA alone and complexed
with thrombin, its specific ligand, by using a network consisting of two different networks. The results are
quite intriguing, confirming some conjectures about the different role of two cations, i.e. Na
+
and K
+
, in
stabilizing the compound. Furthermore, this study suggests the use of resistance measurements to discriminate
among different affinities.
1 INTRODUCTION
The current trend in medicine is the improvement of
prevention (vaccination, disease screening, correct
lifestyle, etc.), the personalization of treatments and a
less invasive and friendly (for example, point-of-care)
diagnostics. Accordingly, the development of new
techniques and therapies is widely explored.
Outstanding results are given by aptamers, which are
small fragments of ssDNA or RNA, artificially
produced to perfectly adapt to an assigned ligand
(from small molecules to large proteins).
The selection and amplification technique used to
produce aptamers is called SELEX (Systematic
Evolution of Ligands by EXponential Enrichment)
(Oliphant, 1989, Ellington, 1990, Tuerk, 1990). This
technique seems so powerful to produce, in principle,
an aptamer for each specific pathogen or
macromolecule found at the origin of a disease. It
could be a revolution in medicine.
In the last 20 years, big efforts have been devoted
to the production of even more efficient aptamers, by
using both biochemical and computational techniques
(Yüce, 2015, Jo, 2016). Some of them are at the
first/second stage of clinical trials, and this result
gives hope of a more and more massive use in
medicine (Ni, 2011).
The mechanism of ligand binding has been
compared to the structural recognition process used
by antibodies to capture antigens, therefore aptamers
are also known as “chemical antibodies” (Sun, 2014).
Like antibodies, they bind the target with high
specificity and selectivity and, therefore, this has
awakened a wide interest for the possible
technological uses. As a consequence, a rapid
development of aptasensors, i.e. sensors based on
aptamers (Iliuk, 2011) has taken place. Indeed,
aptamers are used in biosensors in substitution of
antibodies, which are, usually, more difficult to
produce and often require animal sacrifice. Despite
all the progresses made in the field of aptamer
production and selection, so as in the aptamer
technological and medical applications, the
understanding of the biochemical and physical
processes underling the aptamer/protein-ligand
interaction is still quite poor (Du, 2016).
In this paper we focus attention on the small 15-
mer TBA (5’-GGT TGG TGT GGT TGG-3` ), whose
ability in the inhibition of the enzyme thrombin is
well known. Thrombin is an enzyme present in
62
Alfinito, E., Cataldo, R. and Reggiani, L.
A Network of Networks to Reproduce the Electrical Features of an Aptamer-ligand Complex - What an Electrical Network Tells about Affinity.
DOI: 10.5220/0006361000620069
In Proceedings of the 2nd International Conference on Complexity, Future Information Systems and Risk (COMPLEXIS 2017), pages 62-69
ISBN: 978-989-758-244-8
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

mammals blood where rules coagulation. Therefore,
in some diseases, like heart attack, its inhibition is
part of the therapy. TBA passed the first step of the
clinical trials; at present, a modified version of this
aptamer, which ensures a longer permanence in
human body, has been patented and is currently used
in clinics.
Recently, TBA has been successfully used in a
prototype of thrombin sensor (Cai, 2005). This device
is sensitive to thrombin in a range of 6 orders of
magnitude and selective with respect non-thrombin
molecules. Furthermore, the detection technique,
based on electrochemical impedance spectroscopy, is
well consolidated. At present, the only -although
quite serious- constraint to its large scale diffusion is
miniaturization, which should allow for point-of-care
uses.
The aim of this paper is to show that a network of
networks can be a good tool for investigating some
electrical properties of the complex TBA-thrombin,
each of them represented by a specific network.
Specifically, this tool, which is contained in a more
wide approach called proteotronics (Alfinito, 2015),
is able to correctly describe and interpret some
relevant results obtained by using X-ray spectrometry
(Russo–Krauss, 2012), and electrochemical
impedance spectroscopy (EIS) measurements (Cai,
2005). In particular, this approach is able to foresee
the reduced affinity of the TBA-thrombin composite,
when produced in the presence of Na
+
, with respect
to that of the same compound, produced in a solution
containing K
+
. Furthermore, the resistance variation
observed in EIS measurement is also well
reproduced. Finally, the results reveal a very
interesting feature: the protein binding lowers instead
of increasing the aptamer resistance. As a matter of
fact, compared to the aptamer in the native state, there
is an increase of resistance, in agreement with
experiments, but, compared to the aptamer in the
active state, the resistance decreases. This is an
unexpected result, which is mainly related to the
complex structure of the associated interconnected
networks.
2 MATERIALS AND METHODS
The network of networks is built up starting from the
single networks representing the protein and the
aptamer, respectively. In particular, to build up the
networks we use:
a. the aptamer in its native state, i.e. its lowest
free energy state;
b. The aptamer in its active form, i.e. the
aptamer with the structure deformed due to
the binding but deprived of the protein, in
the presence of both K
+
and Na
+
;
c. The aptamer-enzyme complex, in the
presence of both K
+
and Na
+
;
d. The enzyme alone in the presence of both K
+
and Na
+
.
Structure a. is available from the Protein Data
Bank (Berman, 2000) at the entry 148D (Schultze,
1994), while structures b-d are available from the
Protein Data Bank at the entries: 4DII and 4DIH
(Russo-Krauss, 2012).
Furthermore, each network has been equipped
with specific electrical features and can be used to
explore the topological and electrical properties of the
corresponding biomolecule. The complete network,
the network of networks, is obtained by using the 3D
structure of the complex aptamer-thrombin and
allowing appropriate electrical interactions between
the different macromolecules, as described in Section
2.2
2.1 Topological Methods
Each structure is mapped into a complex network
whose nodes correspond to the position of the C
1
carbon atoms, for the aptamer, and to that of the C
carbon atoms for the enzyme. The nodes are
connected with a simple cut-off rule, i.e. only if the
distance is smaller than an assigned value, R
C.
In such a way, some topological features, like the
structure deformation subsequent to the protein
attachment, can be easily described. Figure 1 shows
the adjacency matrix, also known as contact map
(each point represents the link between the nodes
corresponding to the point coordinates) of the
aptamer in the active form, in the presence of Na
+
and
K
+
. The value of the interaction radius is 10Å.
The contact maps are equal. Nevertheless, the
aptamer-thrombin complex shows tiny differences in
the structure when produced in the presence of the
two different cations, see Figure 2.
In brief, these results tell us that the cations
slightly affect the topological structure of the
complex. Anyway, it is well known that affinity does
depend on the solvent (Russo-Krauss, 2012),
therefore we proceed with a more detailed
investigation.
A Network of Networks to Reproduce the Electrical Features of an Aptamer-ligand Complex - What an Electrical Network Tells about
Affinity
63

Figure 1: Contact map of TBA in the active state, in solution
with two different cations, Na
+
, magenta, and K
+
, black
(color online).
Figure 2: Contact map of the complex TBA-thrombin in
solution with two different cations, Na
+
, black, and K
+
,
green (color online).
Complementary information concerning the network
structure is given by the degree distribution. Figure 3
reports the degree distribution of the TBA in the
native structure and active state, R
C
=10Å.
Figure 3. Degree distribution of TBA in the native and
active state, R
C
=10Ǻ. The degree distribution of the active
state does not depend on the ion in solution (color online).
The degree distribution of the native state has a
typical Poisson-like shape. The degree distribution of
the active state does not depend on the kind of ions
used in the solution; the maximum has shifted at the
value 2, and there are not more nodes with more 5
links. This means that the structure is more dilated
than the native one.
On the other hand, when we give a look to the
degree distribution of the enzyme alone (Figure 4, full
histograms), we notice small but noticeable
differences depending on the used ions.
Figure 4: Degree distribution of the TBA-thrombin
complex, R
C
=10Ǻ (empty histogram) and the thrombin
alone (full histogram).
As expected, in the TBA-thrombin complex
(empty histograms) the number of nodes with a small
number of links is larger (due to the dilated structure
of the aptamer).
The structure resolved in the solution containing
potassium has a Poisson-like degree distribution,
while the structure resolved in a solution containing
sodium is more flat. A more detailed investigation on
this topic is in progress.
In conclusion, the topological analysis reveals that
the differences between the macromolecules in the
presence of the cations K
+
and Na
+
are quite subtle
and mainly concern the degree distribution. These
differences are amplified by the electrical response,
as described in the following section.
2.2 Electrical Methods
To investigate the electrical response of the
macromolecules we built up the corresponding
electrical network.
The single protein electrical network is produced
as in previous investigations (Alfinito, 2009, Alfinito,
2010). In particular, each link is thought as an
electrical line with a specific resistance and
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
64

capacitance. Each passive element of this line is
geometrically represented by a cylinder of height l,
the distance between the nodes, and basis area A, the
intersection area of two spheres of radius R
C
centred
in each of the nodes. The resulting electrical
impedance depends on the kind of nodes (here amino
acids or nucleobases) and on their distance. Finally,
the impedance of the link between the nodes a,b
writes:
Z
a,b
=
l
a,b
A
a,b
ρ
a,b
1+iϱε
0
ε
a,b
ω
(1)
where
a,b
and
a,b
are the resistivity and the
dielectric constant of the link, is the frequency of
the applied voltage. The polarizability values are
quite different for amino acids and nucleic bases (and,
in principle, also the macroscopic electric response of
aptamers could be not the same observed in proteins
(Akimov, 2008)). Here we assume that, in similar
experimental conditions, aptamers and proteins have
similar electrical behaviors and formulate our model
accordingly.
The aptamer electrical features are modelled by
using the values of resistivity and polarizability of the
AGCTU set, recently published (Ohshiro, 2012,
Šponer, 2001). Specifically, the relative dielectric
constant of the couple of the a-th and b-th nodes,
ε
a,b
,
is expressed in terms of the intrinsic polarizability of
each isolated amino acid/nucleobase, α
elec
(Šponer,
2001) , and writes:
,,
,
4
1
6
elect a elect b
ab
(2)
where the second term in the r.h.s. describes the
mean value of the polarizability of the couple of
nodes.
The resistivities of the nucleobases are taken by
(Ohshiro, 2012) ,
aa
, where
is the mean
value calculated upon the AGCTU set and is the
fraction /ρ for each nucleobase.
Therefore, the resistivity of the link drawn between
the a-th and the b-th nucleobase is defined as :
,
.
2
ab
N
ab
(3)
Since analogous data are not given for amino
acids, for the sake of simplicity, we assume that their
resistivities are all the same. In particular, the link
resistance of a couple of amino acids is:
A
a,b
=
. (4)
The region between the networks, where the
hooking happens, is the most interesting. Here the
protein perfectly matches the aptamer and combines
with it via van der Waals forces. It has been revealed
that the presence of some ions like Na
+
and K
+
gives
TBA a different ability to inhibit thrombin (Russo-
Krauss, 2012). Furthermore, in previous sections we
have seen that these cations poorly affect the TBA-
enzyme structure.
Accordingly, we conjecture that most of these
differences are due to the small interaction region.
Therefore, we assume that previous formulas still
hold in each part of the network of networks. This is
also true in the aptamer-thrombin contact region, but
with features intermediate between those of amino
acids and nucleobases. In particular, for the couple of
the a-th nucleobase and b-th amino acid, the link
resistance is:
,
1
.
2
a
NA
ab
(5)
Equation (2) is modified accordingly.
Each electrical network is ideally contacted to an
external bias, by using the first and the last node as
terminals. The electrical features of this linear
network are calculated by using the Kirchhoff laws,
applied to a set of linear equations. Finally, the
equations are numerically solved by a standard
computational procedure which has its roots in the so-
called random resistor network method (De
Arcangelis, 1985, Pennetta, 2004) and has driven to
the proteotronic approach (Alfinito, 2015, Alfinito,
2009).
2.3 Thrombin Inhibition - the Energy
Funnel Framework
The exact mechanism of thrombin inhibition is, at
present, quite unclear and, in general, the protein
activation due to a specific ligand is a long time
debated problem (Onuchic, 1994, Kobilka, 2007).
Here we describe the process of TBA-thrombin
conjugation from the point of view of the energy
transitions the single macromolecule and the
compound perform (Alfinito, 2017, Alfinito, 2016).
First of all, when the protein/aptamer assumes its
native and stable configuration, its energy is on the
bottom of a configurational energy funnel (the native
funnel) which corresponds to the set of energies the
macromolecule has when it goes from the molten to
A Network of Networks to Reproduce the Electrical Features of an Aptamer-ligand Complex - What an Electrical Network Tells about
Affinity
65
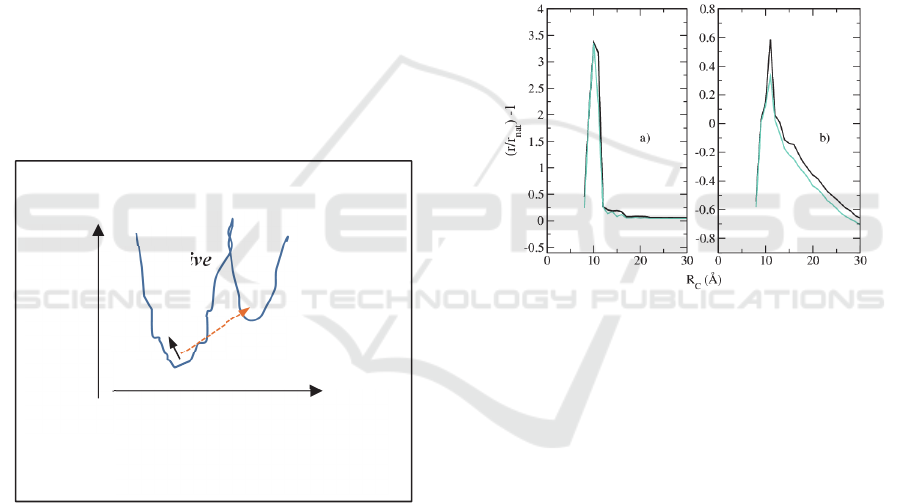
the native state. When it receives energy from the
environment it can go up in the funnel. A way to
receive energy is to be surrounded by the specific
ligands. The ligands can smoothly improve the
aptamer energy simply striking it. Otherwise, when
they bind the aptamers, they produce a sharp energy
jump into a different funnel, the bond funnel. As a
final result, the macromolecule changes its
conformation and binds the ligand. Therefore, the
energy funnel changes (binding funnel). In
conclusion, the addition of specific ligands produces
both the shift of energy in the native funnel and the
transition to the binding energy funnel. A cartoon
describing these two mechanisms is given in Figure
5. This complex mechanism is described, inside the
impedance network analogue, with a change of the
value of R
C
(Alfinito, 2017, Alfinito, 2016). In doing
so, we follow the model proposed by Kobilka and
Deupi (Kobilka, 2007) which describes the protein
dynamics in a process of binding as a transition
between a couple of energy funnels. Furthermore, this
induces a change of the number of internal bonds,
preserving only those useful for stabilizing the final
configuration.
Figure 5: Sketch of the two possible energy transformations
between two contiguous energy funnels, due to the aptmer-
ligand interaction. Continuous line represents a smooth
transition and dashed line an abrupt transition.
3 THEORY AND EXPERIMENTS
In this section we compare some experimental data
concerning the structural and inhibitory properties of
TBA with theoretical data coming from our model.
3.1 Resistance Data
It is well known that ions play an important role in
stabilizing the 3D structure of TBA, the so-called G-
quadruplex. In particular, by adding K
+
ions, the
result is a more stable G-quadruplex and an increased
inhibitory activity of thrombin (Russo- Krauss, 2012)
.
The resistance spectra produced within the
proteotronic approach are represented in Figure (6).
In particular, in Figure 6a, the resistance of TBA
activated in the presence of K
+
and Na
+
is reported in
comparison with the resistance of TBA in the native
state, for different values of R
C
. Activation produces
a resistance increase mainly in the region 8-16 Ǻ. The
main difference is obtained for R
C
=10Ǻ and is the
same for both the activated structures.
Figure 6. Relative resistance variation of TBA in the active
state (a), or complexed with thrombin (b), vs. TBA in the
native state. Turquoise lines refer to structures in the
presence of K
+
, black lines refer to structures in the
presence of Na
+
(color online).
On the other hand, when the protein is added,
Figure 6b, the resistance variation becomes strongly
depend on the kind of ions in solution. Furthermore,
it is quite smaller than that observed for the activated
structures (notice the different scales in Figures (6a)
and (6b)). Therefore, the protein does not produce a
simple passivation, but it integrates the network of the
TBA in the active state and globally reduces its
resistance.
The TBA-thrombin structure obtained in the
presence of K
+
has a larger resistance compared to the
same structure obtained in the presence of Na
+
, more
close to that observed in experiments. This prompt us
to explore this structure for further investigations.
Free
energy
Conformational entropy
native
bond
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
66

3.2 EIS Data: Experiment
The experimental outcomes we refer for testing our
model concern with the use of TBA in an
electrochemical impedance spectroscopy assay (Cai,
2005).
In particular, a gold electrode, functionalized with
TBA, was used to perform electrochemical
measurements in the absence of thrombin and after
incubation with different concentrations of the
enzyme (from to 1 pM to 1 M). The increasing
protein concentration produced an increase of the
electron transfer resistance, sufficient to fairly resolve
different concentrations (a variation larger than
150 % for the highest concentration was measured).
A sketch of the experiment is shown in Figure 7.
Figure 7. Schematic view of the experiment by Cai and co-
workers. On top, the gold electrode functionalized with
TBA without (left) and with (right) thrombin. On bottom
the EIS response, with a sensitive variation of impedance
due to thrombin.
3.3 EIS Measurements: Theory
Our model describes the behaviour of a single
macromolecule, therefore, to compare our data with
experiments, it is necessary to perform a rescaling. In
particular, we are interested in the comparison of
resistance data and it is made by using the following
formula (Alfinito, 2017):
sample com nat
rNfr(1f)r
(6)
where r
sample
is
the sample resistance, r
com
the
resistance of a single TBA-thrombin complex, and
r
nat
the resistance of the single TBA in the native state. N
is the total number of aptamers on the electrode and f
is the fraction of them bound to the protein. We
estimate the f value by assuming it is described by the
Hill-like equation:
a
a
x
f
bx
(7)
where
0C0
)/RR(R x
, R
0
is the value of R
C
corresponding to f=0 i.e. a sample consisting only of
TBAs in their native state, and a and b are numerical
fitting parameters. Equation (7) states a functional
dependence between f and the value of the interaction
radius, in agreement with Sec.2.3.
In the present case, we assume R
0
=13.3Å, which
is the value corresponding to zero difference between
the resistance of the aptamer alone and that of the
complete macromolecule. Furthermore, this value is
the largest corresponding to the region in which, in
agreement with experiments, the resistance of the
complex TBA-thrombin is larger than that of the TBA
in the native state (see Figure 6). By simultaneously
using Equations (6,7) we are able to reproduce the
experimental data. In particular, by using in Equation
(7) the fitting parameters, a=2.99 and b=2.7 10
-4
,
Equation (6) reproduces the sample resistance
variation observed by Cai and coworkers.
The results are given in Table 1.
Table 1: The rate r/r
0
of the sample electron-transfer
resistance, R
et
, measured for different thrombin
concentrations with respect the sample R
et
without
thrombin; the corresponding theoretical quantities
calculated for the values of R
C
shown in column 3,
and the
fraction, f, of aptamer-thrombin complex.
r/r
0
experiment
r/r
0
theory
R
C
(Å)
f
2.6±0.6 3.2 11.3 0.93
2.2±0.5 2.1 11.5 0.90
2.2±0.7 1.8 11.7 0.87
1.7±0.5 1.2 12.7 0.26
4 CONCLUSIONS
We have used a network of networks for analysing
the electrical features of the complex constituted by
the aptamer TBA and its specific ligand, the enzyme
thrombin. The inhibition activity of TBA on this
protein is a long time known result, also investigated
to produce a targeted therapy with reduced side
-
I
mZ
-
I
mZ
R
eZ
R
eZ
A Network of Networks to Reproduce the Electrical Features of an Aptamer-ligand Complex - What an Electrical Network Tells about
Affinity
67

effects. Furthermore, it has been also considered for
producing a thrombin biosensor.
We have adapted a procedure previously used for
a single protein to the aptamer alone, complexed with
the specific ligand, and also without the ligand but
with the modified structure it assumes when binds the
ligand. The aptamer structures, taken by the Protein
Data Bank, describe the oligomers in two different
solutions. We observe that the electrical responses of
the corresponding networks do not depend on the
kind of solution (with Na
+
or K
+
), for the case of the
aptamer alone. On the other hand, when the protein
binds the aptamer, the different action of the two
cations is reflected by a different resistance response.
Thus, this definitely confirms the relevant role of the
cations in the binding mechanism. In other words, the
cation steric action determines the shape of the
network, and finally, the inhibition activity of TBA.
In a more pragmatic approach this results suggest that
a measure of resistance could be a test of affinity.
Another important result obtained with the
technique of the network of networks is that by
adding a large protein like the thrombin to TBA in its
active form, the global resistance is lower than that of
the aptamer. This is an important information
concerning the mechanism of binding because it
reveals that the protein efficaciously completes the
not trivial structure of the aptamer, producing a global
improvement of its conductance. Of course, and in
agreement with experiments, the final resistance
value is lower than that of the aptamer in the native
state, but larger than that of the aptamer in the active
state.
This enforces the conclusion that, at this level of
microscopic interactions, the bulk approximation
fails.
REFERENCES
Akimov, V., Alfinito, E., Bausells, J., Benilova, I., Paramo,
I.C., Errachid, A., Ferrari, G., Fumagalli, L., Gomila,
G., Grosclaude, J. and Hou, Y., 2008. Nanobiosensors
based on individual olfactory receptors. Analog
Integrated Circuits and Signal Processing, 57(3),
pp.197-203.
Alfinito, E., Pennetta, C. and Reggiani, L., 2009.
Topological change and impedance spectrum of rat
olfactory receptor I7: A comparative analysis with
bovine rhodopsin and bacteriorhodopsin. Journal of
Applied Physics, 105(8), p.084703.
Alfinito, E., Pennetta, C. and Reggiani, L., 2010. Olfactory
receptor-based smell nanobiosensors: an overview of
theoretical and experimental results. Sensors and
Actuators B: Chemical, 146(2), pp.554-558.
Alfinito E., Pousset J., and Reggiani L.,2015 Proteotronics:
Development of Protein-Based Electronics. CRC Press.
Alfinito, E. and Reggiani, L., 2016. Current-voltage
characteristics of seven-helix proteins from a cubic
array of amino acids. Physical Review E, 93(6),
p.062401.
Alfinito, E., Reggiani, L., Cataldo, R., De Nunzio, G.,
Giotta, L. and Guascito, M.R., 2017. Modeling the
microscopic electrical properties of thrombin binding
aptamer (TBA) for label-free biosensors.
Nanotechnology, 28(6), p.065502.
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat,
T.N., Weissig, H., Shindyalov, I.N. and Bourne, P.E.,
2000. The protein data bank. Nucleic acids research,
28(1), pp.235-242.
De Arcangelis, L., Redner, S. and Coniglio, A., 1985.
Anomalous voltage distribution of random resistor
networks and a new model for the backbone at the
percolation threshold. Physical Review B, 31(7),
p.4725.
Du, X., Li, Y., Xia, Y. L., Ai, S. M., Liang, J., Sang, P., Ji,
X.-L. and Liu, S. Q., 2016. Insights into Protein–Ligand
interactions: mechanisms, models, and methods.
International journal of molecular sciences, 17(2),
pp.144-177.
Iliuk, A.B., Hu, L. and Tao, W.A., 2011. Aptamer in
bioanalytical applications. Analytical chemistry.
83(12), pp. 4440-52.
Ellington, A.D. and Szostak, J.W., 1990. In vitro selection
of RNA molecules that bind specific ligands. Nature,
346, pp.818–822.
Jo, H. and Ban, C., 2016. Aptamer–nanoparticle complexes
as powerful diagnostic and therapeutic tools.
Experimental & Molecular Medicine. 48(5). e230.
Kobilka, B.K. and Deupi, X., 2007. Conformational
complexity of G-protein-coupled receptors. Trends in
pharmacological sciences, 28(8), pp.397-406.
Ni, X., Castanares, M., Mukherjee, A., Lupold, SE., 2011.
Nucleic acid aptamers: clinical applications and
promising new horizons. Current medicinal chemistry.
18(27), p.4206.
Ohshiro, T., Matsubara, K., Tsutsui, M., Furuhashi, M.,
Taniguchi, M. and Kawai, T., 2012. Single-molecule
electrical random resequencing of DNA and RNA.
Scientific reports, 2, p.501.
Oliphant, AR, Brandl, C.J. and Struhl, K., 1989. Defining
the sequence specificity of DNA-binding proteins by
selecting binding sites from random-sequence
oligonucleotides: analysis of yeast GCN4 proteins.
Mol. Cell. Biol. 9, pp. 2944–2949.
Onuchic, J.N., Luthey-Schulten, Z. and Wolynes, P.G.,
1997. Theory of protein folding: the energy landscape
perspective. Annual review of physical chemistry,
48(1), pp.545-600.
Pennetta, C., Alfinito, E., Reggiani, L., Fantini, F.,
DeMunari, I. and Scorzoni, A., 2004. Biased resistor
network model for electromigration failure and related
phenomena in metallic lines. Physical Review B,
70(17), p.174305.
COMPLEXIS 2017 - 2nd International Conference on Complexity, Future Information Systems and Risk
68

Russo-Krauss, I., Merlino, A., Randazzo, A., Novellino, E.,
Mazzarella, L. and Sica, F.,2012. High-resolution
structures of two complexes between thrombin and
thrombin-binding aptamer shed light on the role of
cations in the aptamer inhibitory activity. Nucleic Acids
Research. 40, gks 512.
Schultze, P., Macaya, R.F. and Feigon, J., 1994. Three-
dimension al solution structure of the thrombin-binding
DNA aptamer d (GGTTGGTGTGGTTGG). Journal of
molecular biology. 235, pp. 1532-1547.
Šponer, J., Leszczynski, J. and Hobza, P ., 2001. Electronic
Properties, Hydrogen Bonding, Stacking,and Cation
Binding of DNA and RNA Bases. Nucleic Acid Sci.,
(61), pp.3–31.
Sun, H., Zhu, X., Lu, P. Y., Rosato, R.R., Tan, W. and Zu,
Y., 2014. Oligonucleotide aptamers: new tools for
targeted cancer therapy, Molecular Therapy-Nucleic
Acids 3, e182.
Tuerk, C. and Gold, L., 1990. Systematic evolution of
ligands by exponential enrichment: RNA ligands to
bacteriophage T4 DNA polymerase. Science. 249,
pp.505–510.
Yüce M., Ullah N. and Budak H., 2015. Trends in aptamer
selection methods and applications. Analyst., 140(16) ,
pp.5379-99
A Network of Networks to Reproduce the Electrical Features of an Aptamer-ligand Complex - What an Electrical Network Tells about
Affinity
69
