
A Semantic Web Engine for Biorefining Model Integration
Edlira Kalemi, Linsey Koo and Franjo Cecelja
Centre for Process & Information Systems Engineering, University of Surrey, Guildford GU2 7XH, U.K.
Keywords: Semantic Engine, Ontologies, OWL-S Services, Biorefining Model Integration.
Abstract: The number of models available to the biorefining community is continuously increasing, there is a need to
provide better ways for their description, categorization, discovery and integration in order to improve
reusability of them. Biorefining models on the other hand are developed using heterogeneous methods, data
format and various environments that makes their reuse challenging. In this paper, we describe a semantic
web engine for the domain of biorefining, which enhances the description of biorefining model by using
semantic web technologies in order to facilitate discovery and integration. In particular, we present how
domain and web service ontologies are used in semantic mapping for the purpose of model integration,
which is achieved by OWL-S (Semantic Markup for Web Services). Whilst the application has been
designed and implemented for a specific domain, this novel design can be applicable to similar problems in
other domains.
1 INTRODUCTION
Mathematical modelling and simulation have been
widely and for long used to represent many aspects
of biorefining i.e. bioenergy production systems,
conversion processes, supply logistics and
environmental impacts (L. Wanga et al., 2015).
Those models can provide powerful tools to design a
biorefining system and evaluate its technical
feasibility, economics and environmental impacts.
As a result, there are a great number of models that
can be shared with the biorefining community and
hence reused. Model integration in biorefining is an
important function, which enables model integration
from unit composition to model orchestration and
invocation. The highest level of integration is the
supply chain integrating lower scale models in
varieties of ways. To this end, biorefining models
are characterized by heterogeneous methods, data
format and different development environments
making their reuse a challenge.
To address this problem, we introduce a semantic
application to enable model sharing, discovery and
integration. Main functionalities of the application
include; i) Model Registration where detailed
information on model functionality, inputs, outputs
and condition of operation are acquired, ii) Model
Publishing which gives the possibility for models to
be shared, iii) Model Discovery which supports the
C6Fermentation">based on different semantic
techniques and iv) Model Composition where the
user has the possibility to integrate his model with
other biorefining models that are discovered through
the application for achieving a more complex goal in
his modelling. Models represent at abstraction level
a biorefining process and are described semantically
by their functionality, inputs, outputs and
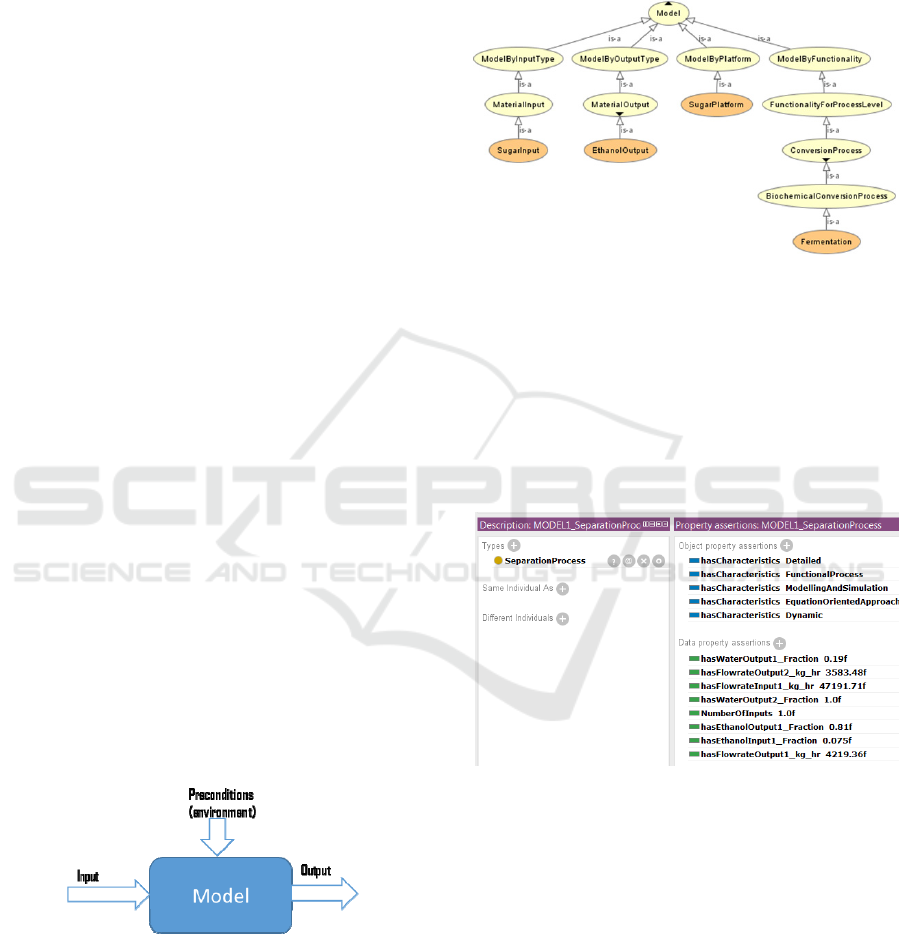
preconditions needed for their execution (Figure 1).
The semantic description of biorefining models is
supported by a biorefining domain ontology, as
demonstrated in the following section. CAPE-OPEN
initiative, a widely recognized industry standard that
defines rules and interfaces in chemical and process
systems community, is the only one that has given
significant contribution in model reusability and
interoperability (Belaud and Pons, 2002) of
Computer-Aided Process Engineering (CAPE)
applications or components. CAPE-OPEN
interfaces, provide the opportunity that process
modelling components can be assembled in process
modelling environments at runtime and has a well-
established communication mechanism by which
software components can exchange functionality at
runtime (Braunschweig et al., 2004). Beside the
advantages that CAPE-OPEN offers, the semantics
of interface are implicit and only human providers
and users are able to decide which software
component will best fit for a specific application's
272
Kalemi, E., Koo, L. and Cecelja, F.
A Semantic Web Engine for Biorefining Model Integration.
DOI: 10.5220/0006439902720279
In Proceedings of the 7th International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH 2017), pages 272-279
ISBN: 978-989-758-265-3
Copyright © 2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
need and therefore the task of identifying the most
adequate model from the libraries is a manual
process (Braunschweig et al., 2004).
2 ONTOLOGY- DRIVEN
APPROACH FOR SEMANTIC
PUBLISHING AND DISCOVERY
Ontologies allow us to define concepts and relations
in the domain of interest, which can be processed by
a machine. OWL (Web Ontology Language) is a
logic based ontology language, which means the
semantics of the language is precisely defined by a
logic. Furthermore, OWL is rich of computation
properties and it is possible to perform computations
automatically using tools, called reasoners (Horridge
et al., 2014). The latest standard ontology languages
is OWL2 (Motik et al., 2009). Reasoners are
software systems that can be used for i) ontology
design time to check certain desirable consequences
follow from what has been stated and ii) application
runtime to provide query answering capabilities,
which ensures queries, such as representing
knowledge is correctly answered. There are many
reasoners, such as FaCT++ (Tsarkov and Horrocks,
2006) and HermiT (Shearer, 2008), that work well
with OWL. A biorefining domain ontology (Koo et
al., 2017) is exclusively developed to describe
biorefining models for the purpose of i) registering
and publishing them based on their semantic
description, ii) performing the process of semantic
discovery, and iii) facilitating the process of
integration. The models in the biorefining domain
ontology are describes in term of functionality,
inputs required for the successful execution of a
model, outputs generated after execution, and the
preconditions of the model, as shown in Figure 1.
Figure 1: Representation of biorefining models.
The ontology is developed in OWL and is often
enriched by adding rules using SWRL (Semantic
Web Rule Language) to provide an additional layer
of expression where OWL lacks expressive power.
The biorefining ontology contains all necessary
concepts and relations for capturing knowledge from
the biorefining domain with a special focus towards
integration of models. Model class is used to collect
information about a biorefining model as an instance
based on functionality, input, output, and biorefining
platform (Figure 2).
Figure 2: Illustration of the hierarchy of classes in the
biorefining domain ontology.
Object and datatype properties are used to define
data inputs and outputs of a model, object properties
are used to further characterize a model and SWRL
rules to define the preconditions which in most cases
in relation to the environment where a model run.
Figure 3 demonstrates an instance of a model in
ontology that belongs to SeparationProcess class.
Figure 3: Separation Process biorefining model instance.
2.1 Registration and Publishing
For publishing and sharing a model, the users will
initially have to register the model by accessing the
semantic web application. In technical terms the
ontology is parsed and checked for consistency by
Hermit reasoner and all classes are given to the user
in the form of a tree (Figure 4a). Figures 4a-c
provides a reduced view of the platform to
demonstrate the operation of the repository to
support the process of registration of the biorefining
models and data.
A Semantic Web Engine for Biorefining Model Integration
273
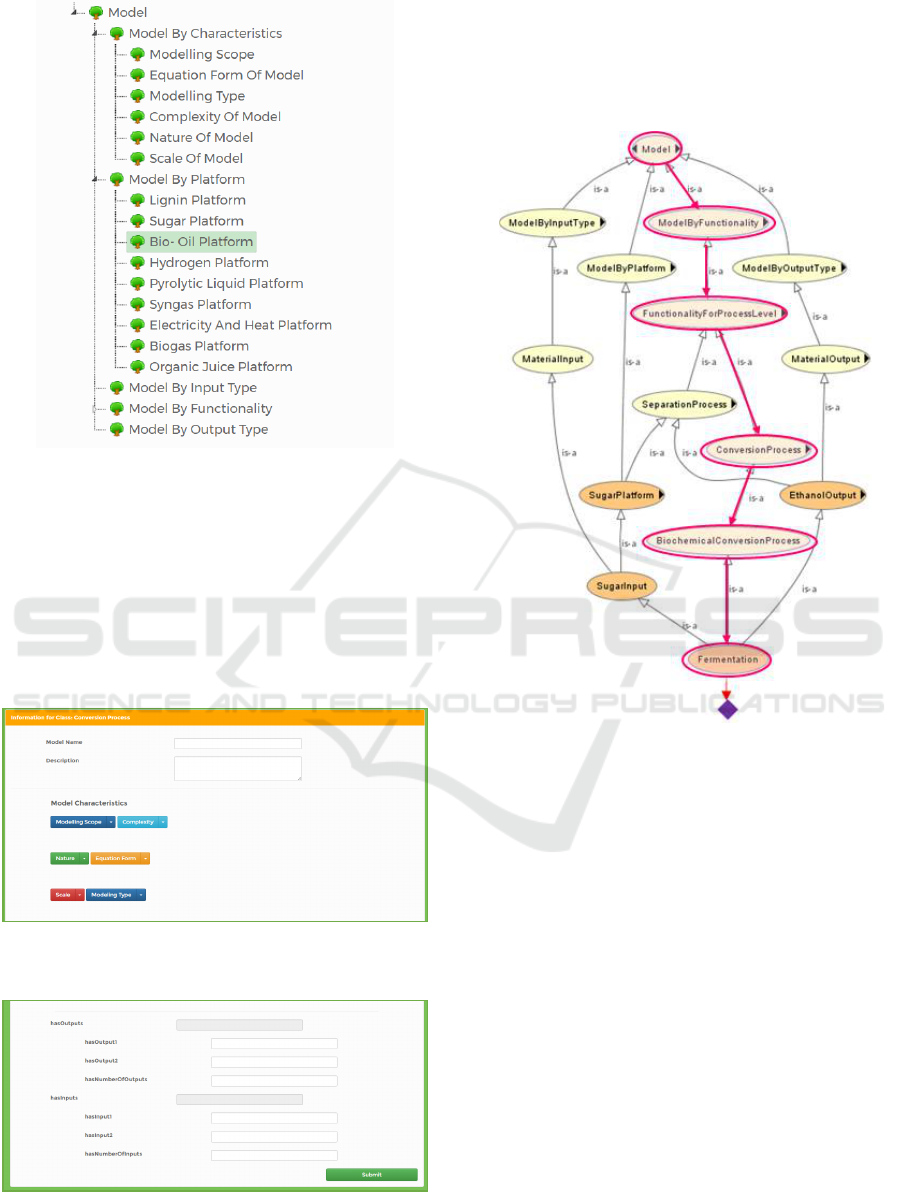
Figure 4a: (Step 1) Model classification.
In step 1 (Figure 4a), user register a model under a
predefined category by browsing a taxonomy
presented in a form of tree. Every class in the
ontology is correlated with a number of datatype and
object properties, mainly in terms of its functionality
input, output and precondition, which is presented in
Figure 4b and c to complete the model registration
process.
Figure 4b: (Step 2) Name and description the model with
annotations and object properties.
Figure 4c: (Step 3) Complete inputs and outputs of the
model by giving value to the data properties.
Illustration of the registration process under
ModelbyFunctionality category in the biorefining
domain ontology is given in Figure 5. Together with
reasoner, fermentation model is registered in terms
of platform, as well as input and output type.
Figure 5: Registration of a fermentation model in the
biorefining domain ontology.
2.2 Discovery
By registering and publishing the models with the
necessary semantic annotation, biorefining models
are stored in a semantic repository and many experts
in the field can make use of them. Next important
step is to discover the models that the semantic
repository maintains. For discovering the required
model the user can choose among four options:
i) List of all models by different categories:
Browse the models as they are listed
based on different categories (i.e. by
functionality, by output etc.)
ii) Search for the desired model using two
filters: a) functionality and b) output of
the model.
iii) Write a SPARQL query to filter the
required models and run it through the
application.
SIMULTECH 2017 - 7th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
274

iv) Semantic matching between a set of
requirements, which is set by the input
of a requesting model and the list of
models in the semantic repository as
candidate models that meet the
requirements. This method is developed
with an intention to achieve the purpose
of model integration and is further
detailed in Section 2.3.
The first and the second techniques are keyword
based queries. The SPARQL editor is made
available by employing jOWL (Decraene, 2007), a
jQuery plugin for OWL-RDFS documents and is
intended to be used by users who are familiar with
knowledge based systems and SPARQL querying
language.
2.3 Semantic Matching
Discovery of candidate models that fully or partially
matches the requirements of a requesting model is a
very important step for a successful model
integration. The requirements of a requesting model
are a set of inputs that the requesting model needs
which has to correspond to the outputs that a model
in the semantic repository has in order to be a
candidate model for integration. This way the
process of matching will result in the process of
comparing the input requirements that the requesting
model has with the outputs that candidate models
offer.
Figure 6: Model input/ output matching.
In our work we have used input/output matching
approach introduced by Cecelja et al. (2015) that
undergoes three-stage matching phases: i)
elimination, where all models that do not satisfy the
critical criteria are excluded from the selection list,
ii) semantic matching by calculating similarity
measures which defines the level of compatibility
between the requested model and the candidate
models and iii) ranking of the candidate models by
similarity measure. The critical criteria is defined by
the user during the formulation of the requesting
model. Semantic matching process calculates the
semantic relevance between the requesting model
and the models published in the repository. This is
calculated as an aggregated value (Algorithm 1) of
distance measure (Algorithm 2) and property
similarity (Koo et al., 2017). Distance measure
signifies the semantic similarity where the ontology
is processed as a graph structure and is calculated by
finding the shortest distance between concepts in the
ontology where paths has different weights. The
quantified distance measure is normalized with the
longest path between concepts in the ontology.
Property similarity is calculated by the average of
cosine and Euclidian similarity using the values of
the properties characterising the models as a vector.
Algorithm 1: Semantic Matching.
Input: O an ontology, M
r
the requesting
model which is instance of O
Output: S, a set of models ordered
based on the semantic matching
For all models in the repository
Eliminate (O, M
r
)
For all models in O after
elemination
CalculateSemanticSimilarity(O[M
i
], M
r
)
CalculatePropertySimilarity(O[M
i
], M)
S[i]←DefineMatchingMatching(Ss,
Qm)
Print S in descanted order
Algorithm 2: CalculateDistanceMeasure.
Input: O an ontology with the
reduces number of instances after the
elimination, the requesting model M
r
Output: K, a number representing the
semantic matching
For all models in the repository
findShortestPath(O, M
r
)
findLongestPath(O)
CalculateK(findShortestPath(O,
M
r
),findLongestPath(O))
Print K
3 SEMANTIC WEB SERVICE
FOR INTEGRATION
One main reason of reusing models in biorefining is
to generate new knowledge from the integration of
heterogeneous data, methods and tools. Semantic
web technologies are considered as the best method
to the support model integration as they use
graphical method to describe data models that
handle unstructured data. The approach we propose
to achieve the model integration in biorefining is
adopted form the semantic web services integration
A Semantic Web Engine for Biorefining Model Integration
275
methodology and the input/output matching. We
consider each biorefining model is an instance of the
domain ontology as a web service and semantically
described with the OWL-S. OWL-S is a well-
established framework with respective service
ontology which, in this case, facilitates a full
automation of model integration. In practical terms,
OWL-S framework (Martin et al. 2004) is
implemented as an ontology with three
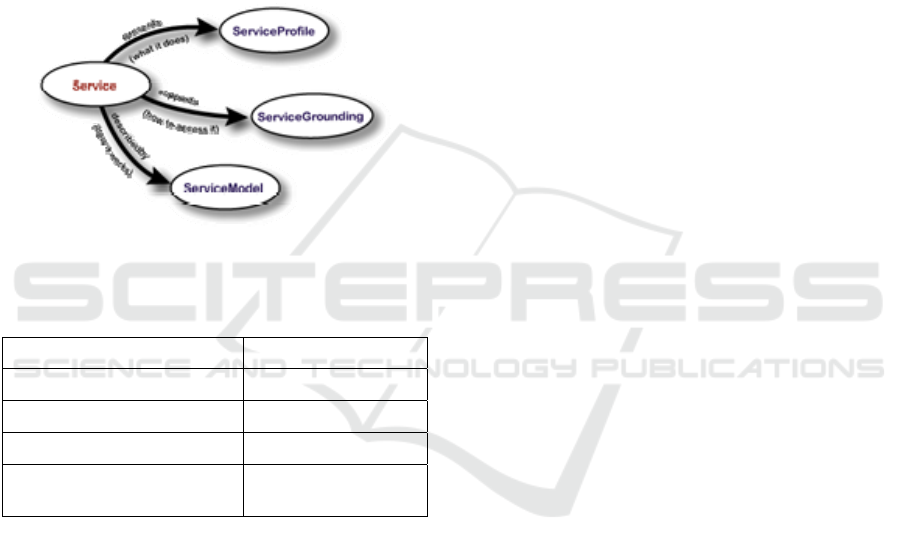
interconnected sub-ontologies (Figure 7); Service
Profile (Profile.owl), Process Model (Proces.owl),
and Process Grounding (Service.owl). Typically a
semantic web service has interlink to all the .owl
files in Table 1.
Figure 7: OWL-S upper layer ontology (Martin et al.
2004).
Table 1: C6Fermentation model as OWL-S service.
C6FermentationService.owl Service instance
C6FermentationProfile.owl Profile
C6FermentationProcess.owl Process model
Grounding.owl Grounding instances
Grounding.wsdl WSDL definitions for
grounding
Within the OWL-S framework, Service Profile
delivers a way to describe services, in our case,
models (Martin et al., 2004) and contains two
different types of information i) contact information
of the model provider and ii) functional description
of the model in terms of inputs required by the
requesting model, outputs generated from the
candidate model, preconditions for execution and
expected effects from the execution (IOPE). Below
is part of the content of the service and profile .owl
files of the C6Fermentation model described by
OWL-S. By taking advantage of OWL-S profiles
structure and their references to OWL concepts of
the biorefining domain ontology, the discovery
process find those models that are most likely to
satisfy the needs of a requesting model (Martin et
al., 2007).
C6FermentationService.owl
…………
<service:Service
rdf:ID="C6Fermentation">
<!-- Reference to the C6Fermentation
Profile -->
<service:presents rdf:resource="URI
C6FermentationProfile.owl#Profile_C6Fer
mentation "/>
<!-- Reference to the C6Fermentation
Process Model -->
<service:describedBy
rdf:resource="URI\C6FermentationProcess
.owl#C6Fermentation_Process"/>………………
</service:Service>
C6FermntationProfile.owl
…………
<!-- reference to the service
specification -->
<service:presentedBy
rdf:resource="URIC6FermentationService.
owl#C6Fermentation "/>
<!-- reference to the process model
specification -->
<profile:has_process
rdf:resource="URIC6FermentationProcess.
owl#C6Fermentation_Process"/>
<profile:serviceName>C6Fermentationt</p
rofile:serviceName>………………………………….
<profile:contactInformation>
<actor:Actor rdf:ID="C6Fermentation ">
<actor:name>C6Fermentation Process
</actor:name>
<actor:phone>67668788</actor:phone>
<actor:fax>412 268 5569</actor:fax>
<actor:email>email@domain.com
</actor:email>
……………………………….
<!-- Descriptions of IOPEs -->
<profile:hasInput
rdf:resource="URIC6FermentationProcess.
owl#TotalFlow"/>
<profile:hasInput
rdf:resource="URIC6FermentationProcess.
owl#Temperature"/>
<profile:hasInput
rdf:resource="URI/C6FermentationProcess
.owl#Pressure"/>
………………………………………………..
<profile:hasOutput
rdf:resource="URI/C6FermentationProcess
.owl#ComponentFractionEthanol"/>…………………
The Process Model describes a model in the sense of
how it works, the tasks it performs, the sequencing
of this tasks, the intermediate inputs and outputs and
the transformation that happens in each stage. A part
of the C6FermentationProcess.owl file code is
presented below, which expresses the process
model.
SIMULTECH 2017 - 7th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
276
C6FermentationProcess.owl
…………………
<process:AtomicProcess
rdf:ID="C6Fermentation">
<rdfs:comment>
Get inputs for successfully running the
model.
</rdfs:comment>
<process:hasInput>
<process:Input rdf:ID="Get_Input_S1">
<process:parameterType
rdf:datatype="http://www.w3.org/2001/XM
LSchema#anyURI">
anyURI/BiorefiningConcepts.owl#ModelFun
ctionality </process:parameterType>
</process:Input>
</process:hasInput>
………………………
<process:hasOutput>
</process:parameterType>
</process:Output>
</process:hasOutput>……….
<process:inCondition>
<expr:SWRL-Condition>
<rdfs:label>hasPlatform</rdfs:label>
<expr:expressionObject>
<swrl:AtomList>
<rdf:first>
<swrl:IndividualPropertyAtom>
<swrl:propertyPredicate
rdf:resource="#hasPlatform"/>
<swrl:argument1 rdf:resource="#GAMS"/>
</swrl:AtomList>
</expr:expressionObject>
</expr:SWRL-Condition>
</process:inCondition>
OWL-S Grounding contain all necessary details for
invoking a model (Pedrinaci et al., 2011) and the
main function is to show how the inputs and outputs
of process are defined concretely as messages,
which carry those inputs and outputs in some
specific transmittable format. To achieve this
purpose OWL-S makes use of the Web Services
Description Language (WSDL) which is a well-
defined language for the purpose.
The model integration phase in the application
starts at the stage of model discovery. The user owns
a model (the requesting model) and tries to discover
other models that could be integrated with his
model, such as models that fully or partially match
the requirement of the model. This phase is
supported by input/output matching approach
explained in section 2.3. The user is given a rank of
available models that are matching fully or partially
to the requesting model. Based on the criteria, user
then selects one of the candidate models.
Automation of the candidate model selection and
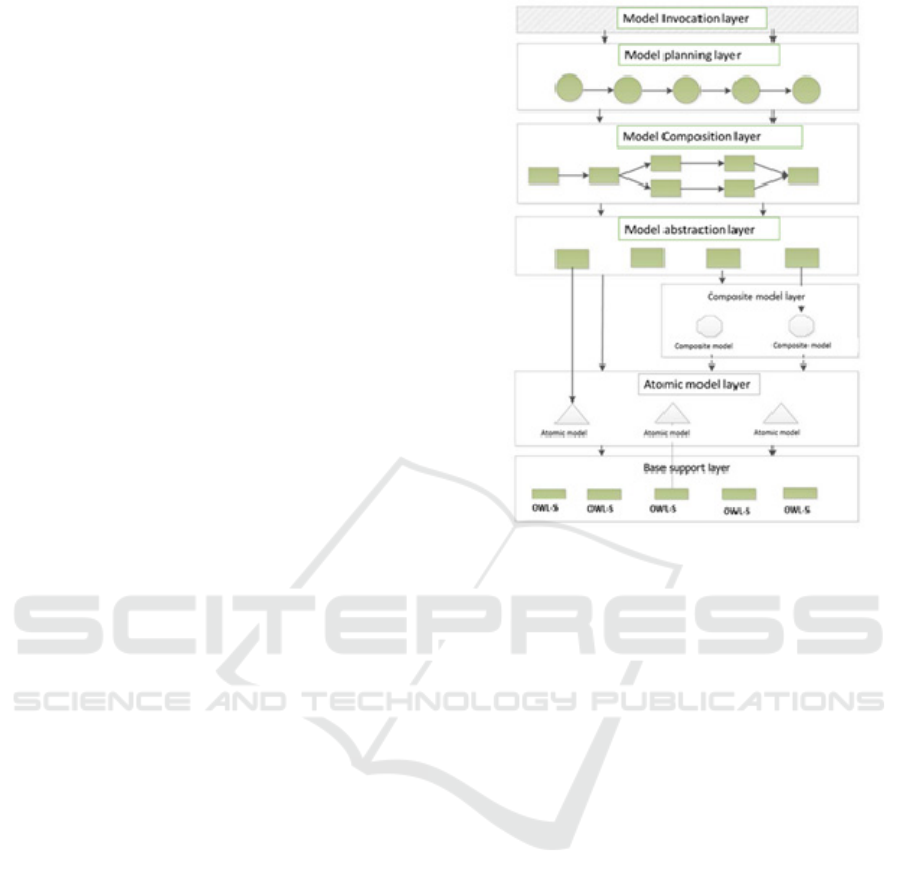
Figure 8: Layered model composition architecture adopted
from Xiang et al 2015.
composition is avoided to give to the user flexibility
of choosing different pathways. Selection of the
most suitable model will call the composition and
invocation process to apply the model and data
integration. Figure 8 gives the model composition
layered architecture where: i) Base layer handles
model registration and invocation ii) Atomic model
layer contains the so called atomic models (simple
model) and serves as the base for planning and
invocation of models iii) Composite service layer
contains composite models (complex model that can
be further decomposed in atomic models) iv) Service
abstraction layer serves as the basis of model
composition v) Model Composition layer composes
models based on their semantic description vi)
Model planning layer provides the sequence of
models to be invoked based on a planning algorithm
vii) Model Invocation layer invoke models based on
the sequence generated from the previous layer.
4 APPLICATION
IMPLEMENTATION
The application is mainly developed using semantic
technologies but is also supported by other web
technologies. Figure 9 displays the semantic
A Semantic Web Engine for Biorefining Model Integration
277

application architecture which is conceptualized in
three main pillars; i) Back end or Service Layer ii)
Middleware and iii) Front End. At the back end, the
semantic web services and the domain ontology are
stored. The core part of the system is developed in
Java and represents the middleware layer, which is
responsible for i) reasoning and interacting with the
knowledge domain ii) match making engine and iii)
interaction of front end interface and services. The
Front End, being a web application, will be accessed
through any browser and this part of the system has
been developed using Bootstrap and Java Server
Pages technologies. Through this interface the users
can i) search for specific models based on specific
criteria (i.e. the functionality of the model or output
of the model or for a set of criteria) ii) registration
of a model and iii) composition (Figure 10) of the
requesting model with the models found in the
repository (candidate models). As the knowledge
base expands the system is updated in real time with
the new knowledge and services. At the middleware
layer a key role plays OWLAPI (Horridge and
Bechhofer, 2011), a Java library for manipulating
OWL ontologies. In collaboration with the reasoner
(i.e. HermiT) and other supporting Java classes,
OWLAPI, are checking the domain knowledge for
consistency, parsing the inferred ontology for
passing it to further elaboration from the front end,
expanding the knowledge base with new instances of
models or integrated models. As the highest aim of
the repository is model integration the input/output
matchmaking module is a crucial component and a
processor step to our end use, the model integration.
For the development of the match maker OWL-S
Java API for parsing, serializing, validation,
reasoning, and execution services for OWL-S (Sirin
and Parsia, 2004).
Figure 9: Application architecture.
The Semantic matching algorithm makes use of the
JENA library (Dickinson et al., 2004), especially for
measuring distances between concepts
(findShortestPath()). Figure 9 shows the stage of the
application where a list of biorefining models is
offered to the user. The user can decide to test the
integration of his model by choosing any of the
candidate models even if the matching between his
model and the candidate ones are not full matching
(100%). Another instrument that can support the
user in his decision in the “Details” section for
every candidate model, where a justification of the
semantic similarity is given. Justification is given by
emphasising matching and non-matching properties
between the requesting and candidate model and the
semantic relevance that the requesting model has
with each of the candidate models.
Figure 10: Part of the application where a list of candidate
models is presented to the user for proceeding with the
selection and integration steps.
5 CONCLUSIONS
A semantic web engine for supporting the
biorefining community in model sharing and reuse
has been introduced. Semantic web technologies are
used to describe biorefining models semantically in
order to assure model discovery and integration
based on the content, instead of just syntax. All the
models are described by the biorefining domain
ontology and further treated and instances of OWL-
S ontology. Users of the application can be experts
of the biorefining community with different levels of
expertise. The application is a complex one and
build upon advanced technologies but this
complexity is hidden to the user and they can benefit
only from the advantage of it. The datatype
properties, inputs and outputs of a biorefining
model, are given by the owner of the model during
registration stage. In some cases the user might need
to test the integration for different data inputs.
Future work will be focused on designing an
intelligent algorithm that will facilitate the
SIMULTECH 2017 - 7th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
278

integration of biorefining models with dynamic data
properties.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the financial
support by the Marie Curie Initial Training
Networks Program, under the RENESENG project
(FP7-607415).
REFERENCES
Belaud, J.P., Pons, M., 2002. Open software architecture
for process simulation: the current status of CAPE-
OPEN standard, Comput. Aided Chem. Eng., 10 (C),
pp. 847–852
Braunschweig, B., Fraga, E., Guessoum, Z., Marquardt,
W., Nadjemi, O., Paen, D., Pinol1, D., Roux, P. Sama,
S., Serra, M., Stalker, I.,Yang, A., 2004. CAPE web
services: the COGents way, Comput. Aided Chem.
Eng., 18 (C) , pp. 1021–1026
Carroll,J., Dickinson,I., Dollin,C., Reynolds, D., Seaborne,
A., Wilkinson, K. , 2004. Jena: implementing the
semantic web recommendations, 04 Proceedings of the
13th international World Wide Web conference on
Alternate, Pages 74-83, doi>10.1145/1013367.
1013381
Decraene, D. 2007. jOWL. [online] Available at:
http://ontologyonline.org/ [Accessed 20 March 2017].
F. Cecelja, N. Trokanas, T. Raafat,M. Yu, 2015. Semantic
algorithm for Industrial Symbiosis network synthesis.
Computers &Chemical Engineering, Online.
Horridge, M., Bechhofer S., 2011. The OWL API: A Java
API for OWL ontologies. Semant. web 2
Horridge, M., Brandt, S., Parsia, B. and Rector, A. L.,
2014, A Domain Specific Ontology Authoring
Environment for a Clinical Documentation System,
IEEE 27th International Symposium on Computer-
Based Medical Systems, New York, NY, pp. 329-334.
doi: 10.1109/CBMS.2014.84
Koo, L., Trokanas, N., and Cecelja, F. , 2017. A semantic
framework for enabling model integration for
biorefining. Computers & Chemical Engineering.
http://doi.org/10.1016/j.compchemeng.2017.02.004
Martin, D.,. Burstein, M., Lassila, O., Paolucci, M., Payne,
T. and McIlraith, S., 2004. Describing Web Services
using OWL-S and WSDL. http://www.daml.org/
services/owl-s/1.1/owl-s-wsdl.html
Motik, B., Patel-Schneider, P. F., Parsia, B., 2009. OWL 2
Web Ontology Language structural specification and
functional style syntax. W3C - World Wide Web
Consortium, W3C Recommendation, [Online].
Available: http://www.w3.org/TR/owl2-syntax/
Pedrinaci, C., Domingue,J., Sheth, P. A.:Semantic Web
Services. Handbook of Semantic Web Technologies
2011: 977-1035
Shearer, R., Motik, B., Horrocks, I., 2008. HermiT: A
Highly-Efficient OWL Reasoner in Proc. of the 5th
Int. Workshop on OWL: Experiences and Directions
(OWLED 2008 EU)
Sirin, E., Parsia, B., 2004. The OWL-S Java API,
Proceedings of the Third International Semantic Web
Conference
Tsarkov, D., Horrocks, I., 2006. FaCT++ description logic
reasoner: System description in Proc. of the Int. Joint
Conf. on Automated Reasoning (IJCAR 2006),
Lecture Notes in Artificial Intelligence, vol. 4130.
Springer, 2006, pp. 292-297. [Online]. Available:
download/2006/TsHo06a. pdf
Wanga, L., Agyemangb, S. A., Aminib, H., Shahbazia, A.,
Mathematical modeling of production and biorefinery
of energy crops, Renewable and Sustainable Energy
Reviews, Volume 43, March 2015, pp 530-544, ISSN
1364-0321, http://dx.doi.org/10.1016/j.rser.2014.11.
008.
Xiang, D., Xie, N., Ma, B. and Xu, K., 2015.The
Executable Invocation Policy of Web Services
Composition With Petri Net. Data Science Journal, 14:
5, pp. 1-15, DOI: http://dx.doi.org/10.5334/dsj-2015-
005
A Semantic Web Engine for Biorefining Model Integration
279
