
Anomaly Detection for Industrial Big Data
Neil Caithness and David Wallom
Oxford e-Research Centre, Dept. of Engineering Science, University of Oxford, Oxford, U.K.
Keywords:
Industrial Internet of Things (IoT, IIoT), Industrial Big Data, Anomaly Detection, Ordination,
Singular Value Decomposition (SVD), Principal Components Analysis (PCA), Correspondence Analy-
sis (CA).
Abstract:
As the Industrial Internet of Things (IIoT) grows, systems are increasingly being monitored by arrays of
sensors returning time-series data at ever-increasing ‘volume, velocity and variety’ (i.e. Industrial Big Data.
An obvious use for these data is real-time systems condition monitoring and prognostic time to failure analysis
(remaining useful life, RUL). (e.g. See white papers by Senseye.io, Prognostics - The Future of Condition
Monitoring, and output of the NASA Prognostics Center of Excellence (PCoE)). However, as noted by others,
our ability to collect “big data” has greatly surpassed our capability to analyze it. In order to fully utilize
the potential of Industrial Big Data we need data-driven techniques that operate at scales that process models
cannot. Here we present a prototype technique for data-driven anomaly detection to operate at industrial scale.
The method generalizes to application with almost any multivariate data set based on independent ordinations
of repeated (bootstrapped) partitions of the data set and inspection of the joint distribution of ordinal distances.
1 INTRODUCTION
Data from energy smart meters represents an appro-
priate use-case for demonstrating a new method for
detecting anomalies in Industrial Big Data, specifi-
cally where accurate system process models do not
exist and where data-driven techniques are sought in-
stead. The electricity network is a system being mon-
itored by arrays of sensors (smart meters), and repre-
sents appropriately high volume, velocity, and variety
of data (de Mauro et al., 2015).
Energy theft by meter tampering represents a sig-
nificant threat to the UK energy industry. The cre-
ation of new opportunities for cyber theft and fraud
introduced by the imminent deployment of up to 50M
smart meters across the UK industry has prompted a
search for a data-driven solution to detecting possi-
ble theft or fraud events. In the DIET project (Data
Insight against Energy Theft
1
) we developed a new
technique using unsupervised machine learning for
the identification of anomalous events based on data
collected exclusively from smart meters.
Electricity smart meters typically collect standard-
ized consumption data (kWh in 48 half-hour bins per
day) and an associated stream of non-standardized
or meter specific “event” or “logging” data. Event
1
Innovate UK, Grant Reference Number 102510.
streams may consist of nominal codes that track the
actions on the smart meter and may relate to activities
such as accessing the cache via modem, turning the
power supply off, or many more of increasingly tech-
nical nature. It is yet unknown whether meter tam-
pering for theft, fraud, or otherwise, carries with it
specific and detectable event sequence signatures (i.e.
there is no relevant process model in existence.)
The question then becomes, can theft or meter
tampering be credibly detected from the data streams
alone, without reference to defining event sequence
signatures? Without credible and/or sizable training
sets of theft or fraud cases with which to train super-
vised machine learning systems, we position this task
instead in the context of anomaly detection, using un-
supervised machine learning techniques.
This is closely related to the field of outlier detec-
tion in the data science literature, where, in a well-
known definition by Hawkins (1980) an outlier is
an observation which deviates so much from
other observations as to arouse suspicion that
it was generated by a different mechanism.
Or, according to Barnett and Lewis (1994)
an observation [...] which appears to be in-
consistent with the remainder of that set of
data.
Caithness, N. and Wallom, D.
Anomaly Detection for Industrial Big Data.
DOI: 10.5220/0006835502850293
In Proceedings of the 7th International Conference on Data Science, Technology and Applications (DATA 2018), pages 285-293
ISBN: 978-989-758-318-6
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reser ved
285

A resurgence of research activity on outlier detec-
tion algorithms was triggered by the seminal work of
Knorr and Ng (1997), and new models have contin-
ued to be developed (Aggarwal, 2013; Akoglu et al.,
2015; Chandola et al., 2009; Schubert et al., 2014),
though none has achieved universal acclaim as being
applicable and effective in all cases.
Campos et al. (2016) have systematically and thor-
oughly reviewed the literature and development of
unsupervised outlier detection algorithms and found
that “little is known regarding the strengths and weak-
nesses of different standard outlier detection models”,
that “the scarcity of appropriate benchmark datasets
with ground truth annotation is a significant impedi-
ment to the evaluation of outlier methods”, and still
further that “even when labeled datasets are available,
their suitability for the outlier detection task is typi-
cally unknown”. Thus endeavors to develop and crit-
ically evaluate new unsupervised techniques are both
timely and warranted.
We refer to the general concept of outlier instead
as anomaly, to emphasize the suspected origin in a
different underlying mechanism as characterized by
Hawkins (1980) cited above, and to distinguish it
from outliers in the normal tail of a statistical distri-
bution produced by a single unified mechanism. In
this paper we present a new approach to the task of
anomaly detection specifically designed to accommo-
date datasets of different data-types on the same set of
cases (e.g. consumption and event data for a portfolio
of smart meters). We know of no comparable tech-
nique that can be used to perform independent analy-
ses of two data streams and then establish a combined
case-by-case measures of outlierness. Identification
of potential anomalous cases is achieved by inspect-
ing the joint-distribution of distances derived from a
novel density analysis. This joint-distribution is sus-
ceptible to new and special statistical interpretation
that we introduce here, but which we will develop fur-
ther in subsequent publications. Our contention is that
in this application, meter tampering for theft, fraud,
or otherwise, will insert into the data set cases that
are not representative of the background mechanism
of the system as a whole.
Finally, we suggest that this procedure generalizes
to anomaly detection for almost any multivariate data
set with any combination of data-types by applying
independent analyses to two or more (possibly ran-
domized) partitions of the data set and then inspecting
the joint distribution of cases.
2 METHOD
For any system (here the electricity network) being
monitored by technical equipment (here a sensor ar-
ray comprising the set of smart meters) that generates
a multivariate cases-by-variables data set, we devise
the following method of analysis. It is useful here to
refer to a schematic diagram of the workflow of the
method as shown in Figure 1. We describe in detail
the five processes labeled S1-S5.
2.1 Partition (S1)
The first novel aspect of the method is that datasets to
be studied are always split into two or more partitions,
vertically by variables, and each partition is analyzed
separately. Cases need not be present in all partitions
(or a case may have null data across the variables in
a partition) but anomalous cases can be detected only
from the set of cases in common across all partitions.
There are a number of criteria for partitioning:
2.1.1 Different Data Types
The smart meter event/consumption data set presents
time series data that is a mixture of data-types: in-
dividually time-stamped nominal-scale event codes;
and time-binned ratio-scale consumption data. Where
the downstream analysis requires different variants
depending in the data-types of the variables, then par-
titioning on data-type is a natural evolution of the
work-flow.
2.1.2 Random Partitioning
Where partitions do not require different kinds of
analysis we make partitions at random and repeat
many times (a process with similar benefits to statis-
tical bootstrapping (Efron and Tibshirani, 1994)).
2.2 Transform (S2)
There are two steps in the transform process: coding
and ordination.
2.2.1 Coding
For the nominal-scale event data we first transform by
frequency count over some specified time period. We
found by inspection that three months is an effective
period: shorter and many events have zero count, so
this is effectively ignoring potentially relevant data;
longer and the anomalous data produced by the event
of a meter tamper could be so diluted by the back-
ground of normal operation as to go undetected. An
DATA 2018 - 7th International Conference on Data Science, Technology and Applications
286
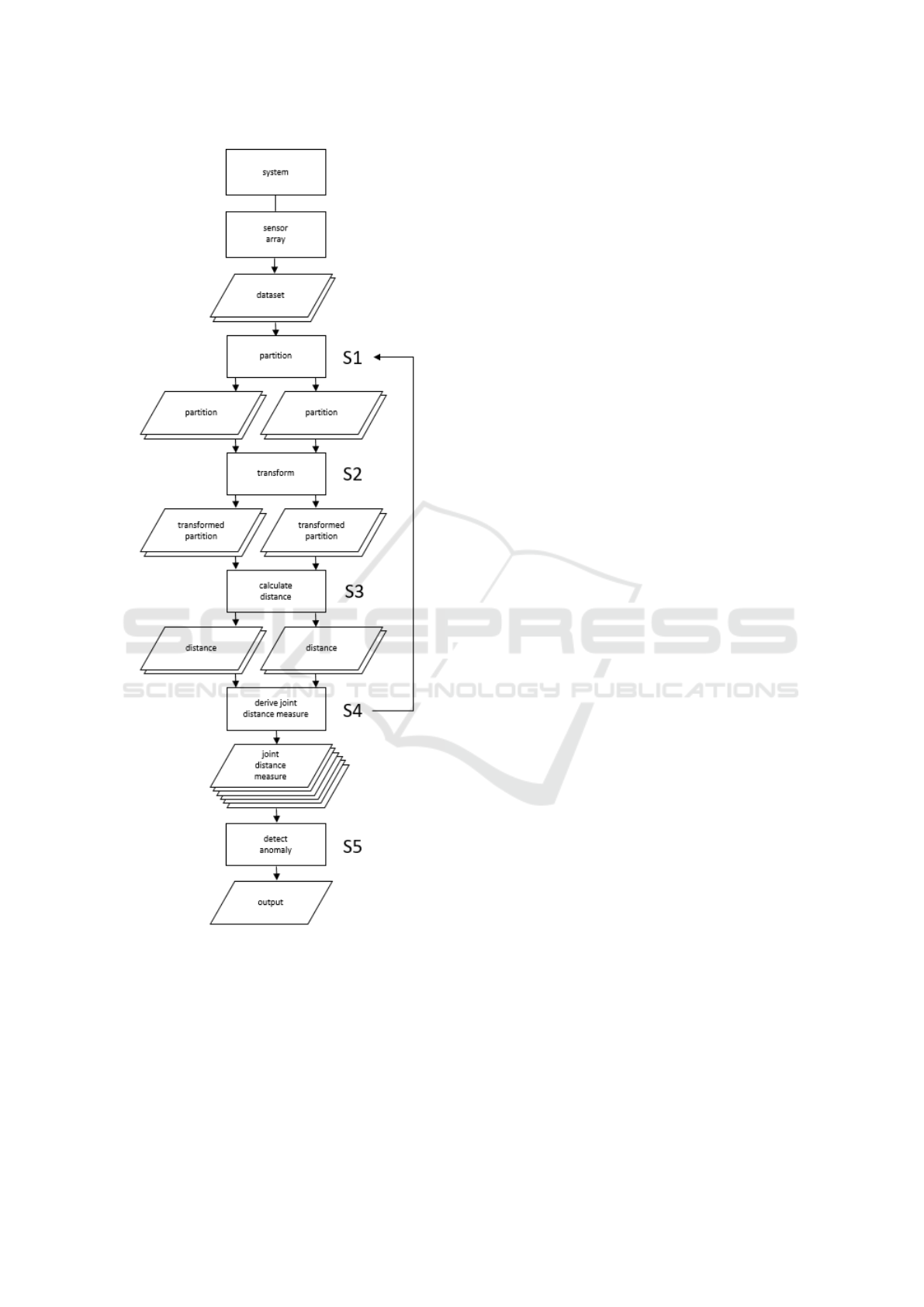
Figure 1: Schematic diagram of the method workflow.
alternative treatment for nominal-scale data would be
to apply a numeric coding. If the partitions are made
at random and repeated sampling is performed, and if
randomized numeric coding is applied with each rep-
etition, then the association between codes remains
unbiased.
For the ratio-scale consumption data, we wish to
capture a relevant cyclical frequency. There are clear
daily, weekly, and seasonal energy consumption pat-
terns, and although these cycles differ between do-
mestic and commercial properties, they remain the
dominant patterns for both types of property. In
our analysis we recode the standard 48 half-hourly
bins per day of consumption data collected by typical
smart meters to a total daily consumption, averaged
by day of the week. We emphasize that this coding
practice was found to be effective in application to
this data set, but is not itself integral to the technique;
there are many other data folding and coding possi-
bilities, and users may find that other transformations
prove effective with their data.
2.2.2 Ordination
We use several variants of methods from the family
of dimension reduction techniques that employ sin-
gular value decomposition (SVD). Specifically, prin-
cipal components analysis (PCA) and correspondence
analysis (CA), depending on the data types of the par-
titions.
The biplot provides a convenient visualization of
the ordination but is not itself integral to the tech-
nique. We illustrate our results with various kinds of
plots, including biplots, in the following section.
The biplot (Greenacre, 2010) is a graphical de-
vice that shows simultaneously the rows and columns
of a data matrix as points and/or vectors in a low-
dimensional Euclidean space, usually just two or
three dimensions. Greenacre (2013) introduced the
contribution biplot in which the right singular vec-
tors (column contribution coordinates) of a dimension
reduction analysis show, by their length, the relative
contribution to the low-dimension solution. Contri-
bution biplots can be used with any of the methods
that perform dimension reduction by singular value
decomposition (SVD), these include correspondence
analysis (CA), principal component analysis (PCA),
log-ratio analysis (LRA), and various derived meth-
ods of discriminant analysis.
SVD is a factorization of a target matrix T such
that
T = UΓV
T
(1)
What distinguishes the various methods is the
form of the normalization applied to T before per-
forming the SVD. In CA this normalization is the ma-
trix of standardized residuals
T = D
−1/2
r
(P − rc
T
)D
−1/2
c
(2)
where P is the co-called correspondence matrix
P = N/n, with N being the original data matrix and
n its grand total, row and column marginal totals of P
Anomaly Detection for Industrial Big Data
287

are r and c respectively, and D
r
and D
c
are the diago-
nal matrices of these.
In the analysis of a cases-by-variables data matrix,
the right singular vectors of the SVD, V, are the con-
tribution coordinates of the columns (variables). A
further transformation involving a scaling factor D
q
,
such that
F = D
−1/2
q
UΓ (3)
defines the principal coordinates of the rows (cases).
The joint display of the two sets of points in F and
V can often be achieved on a common scale, thereby
avoiding the need for arbitrary independent scaling to
make the biplot legible.
The appropriate normalizations and the derivation
of scaling factors for the alternative methods are de-
tailed in his Table 2 and in various equations given
in Greenacre (2013). We use CA for the ordination
of the event data, following a double log transform of
the frequency data, N
0
such that
N = ln(ln(N
0
+ 1) + 1) +1 (4)
Note that the successive additions (+1) in Equa-
tion (4) above are simply to avoid taking ln(0). This is
a convenience, introducing an appropriate scaling so
as to make the biplot legible, but does not otherwise
alter the analysis. For the ratio-scale consumption
data we use the PCA method of Greenacre (2013), af-
ter centering and standardizing the input data by vari-
able.
2.3 Calculate Distance (S3)
Both ordination techniques, whether CA or PCA, re-
sult in a matrix F of principal coordinates of the rows
(cases) as in Equation (3). This matrix has the same
number of dimensions (columns) as variables in the
raw input data, however the information content of
the data is now concentrated towards the higher order
components (i.e. towards the left-most columns of F).
This is the central purpose of the dimension reduction
performed by SVD, and typically, a scree plot is used
to inspect the degree of dimension reduction, essen-
tially a plot of the eigenvalues, Γ in Equation (1).
A decision needs to be made as to how many
components to retain, referred to as a stopping rule
(Jackson, 1993; Peres-Neto et al., 2005). A conven-
tional rule is to retain only those components with
corresponding eigenvalues >1 (known as the Kaiser-
Guttman criterion, (Nunnaly and Bernstein, 1994)),
which is the rule we apply here, though this is a tun-
able parameter of the method and a range of values
should generally be explored. Once a stopping rule
has been decided the case-by-case distances d from
the origin in Euclidean space are calculated for the k
number of retained dimensions. This is done sepa-
rately for each partition. The following code is pro-
vided for clarity.
e.g. Matlab Code
d = sum(F(:,1:k).ˆ2,2).ˆ(1/2)
2.4 Derive Joint Distance Measure (S4)
The second novel aspect of the method, after par-
titioning the data set, is to examine the joint-
distribution of distances derived from the separate or-
dinations of the partitions. Where the partitions have
vastly different numbers and types of variables, and
where the specific ordination techniques differ be-
tween partitions, (as in our case of event data, circa
250 variables of event frequency counts, analyzed
by CA, vs. consumption data, seven variables of
ratio-scale data, analyzed by PCA), then comparison
should be made on the rank order of distances, rather
than directly on the distances themselves.
If all the data across all the variables were gen-
erated by independent random processes, then there
would be no relationship between the rank-ordering
of cases in the two lists. If the variables are at least
partially correlated (as is usually the case for real-
world data) then we would expect a correlation be-
tween the rankings derived from the two partitions,
but we would still expect an even spread of associ-
ations. A scatter plot of the two rank ordered lists
will reveal the nature of the association. A correlation
among variables will manifest as a concentration of
points towards the diagonal, but from a unified under-
lying process we would not expect much departure
from an even spread along the diagonal. If a sec-
ond, distinct process inserts cases into the data set,
we could expect that these may be manifested as a
departure from the uniform density of points, possi-
bly forming locally high density clusters. We plan to
develop the statistics of this phenomenon further in
subsequent publications.
2.5 Detect Anomaly (S5)
Following on from the derivation of a joint distance
measure based on the density of points in the joint
distribution of rank orders, as described in S4, we
now standardize the measure and inspect the depar-
ture from the mean density in units of standard devi-
ation. Cases at the far extremes of departure from the
mean may well be interpreted as being so divorced
from the background process generating the bulk of
the data as to be anomalies produced by a different
mechanism (Hawkins, 1980).
DATA 2018 - 7th International Conference on Data Science, Technology and Applications
288

This final process in the method proceeds to find
those cases at the far extremes of departure from the
mean density, and to report them as likely anomalies
that require an alternative explanation.
3 DATA
We analyzed a data set obtained from British Gas con-
sisting of anonymized records for circa 120,000 smart
meters with daily electricity consumption (kWh in 48
half hour bins) and individually time-stamped events
(for circa 250 different event codes) as logged by each
meter over a three-month period. According to the
data confidentiality agreement we are unable to pro-
vide more specific details or to make these data avail-
able in supplemental material for this publication and
the data set has been returned to ownership of British
Gas. However, we believe the level of detail provided
is sufficient for reproduciblity.
4 RESULTS
We present results of the analysis in a series of plots.
Figure 2 shows a biplot of the consumption data in the
first two dimensions of the PCA ordination. Cases
(the blue dots) can be ranked by their distance from
the origin in any chosen number of dimensions (up to
seven). Interpreting the vectors of the variables (days
of the week) we see the weekend being orthogonal
to week days as we might expect for small business
properties.
The cloud of cases is roughly elliptical in the first
two dimensions with clearly identifiable outliers, but
none that would necessarily arouse suspicion.
Figure 2: PCA biplot of consumption data.
Figure 3 shows a scree plot of the consumption
data ordination. We can be confident that there is
significant dimension reduction (the concave shape).
The Kaiser-Guttman rule selects only the first two
components with eigenvalues >1, however these two
components account for only 45.36% of the variance
of the original data.
Figure 3: Screeplot of consumption data.
Figure 4 shows a histogram of the consumption
PCA ordinal distances with a right-skew, and a not
exceptionally long tail.
Figure 4: Ordinal distance histogram of consumption data.
Figure 5 shows a biplot of the CA ordination of
the event data in the first two dimensions. A hand-full
of variables dominate the solution, in two orthogonal
sets. The rest of the variables contribute only a minor
influence on the solution.
Figure 6 shows a scree plot of the event data or-
dination. Here we can be even more confident that
there is significant dimension reduction. The Kaiser-
Guttman rule (Jackson, 1993) selects about 20 out of
circa 150 variables with eigenvalues >1, and these ac-
count for 88.61% of the variance of the original data.
Figure 7 shows a histogram of the event CA or-
dinal distances with a right-skew, and again a not ex-
ceptionally long tail.
Anomaly Detection for Industrial Big Data
289

Figure 5: CA biplot of event data.
Figure 6: Scree plot of event data.
Figure 7: Ordinal distance histogram of event data.
Figure 8 shows a scatter plot of the ranked dis-
tances from the two ordinations of consumption and
event data. This shows, for the most part, that the data
in the two partitions are independent (the more-or-less
even cloud of data points across the entire space.) If
the variables in the two partitions were partially cor-
related we would expect a concentration in density of
points towards the diagonal.
What we’re not expecting from a unified under-
lying process is (much) variation in density of points
along the diagonal. For these data we see a distinct
cluster of high density at high ranks in the upper right
corner.
Figure 8: Joint rank scatter plot.
Figure 9 shows the density of the joint ranks as a
contour plot where density has been scaled to units
of standard deviation. The high density cluster in the
upper right corner is from two to 40 standard devia-
tions away from the mean of the background process,
and contains no more that 560 cases, out of a total of
some 120k cases. A slight concentration towards the
diagonal is also evident for the background process.
Figure 9: Joint rank density plot.
Figure 10 shows a surface plot using the same
scaling in units of standard deviation. We interpret the
spike in density to indicate a set of anomalous cases
that are probably derived from a different underlying
mechanism to the rest of the dataset.
Figure 11 illustrates the long tail of the distribu-
tion and shows just 560 cases with std >2, out of a
dataset of some 120k cases.
DATA 2018 - 7th International Conference on Data Science, Technology and Applications
290
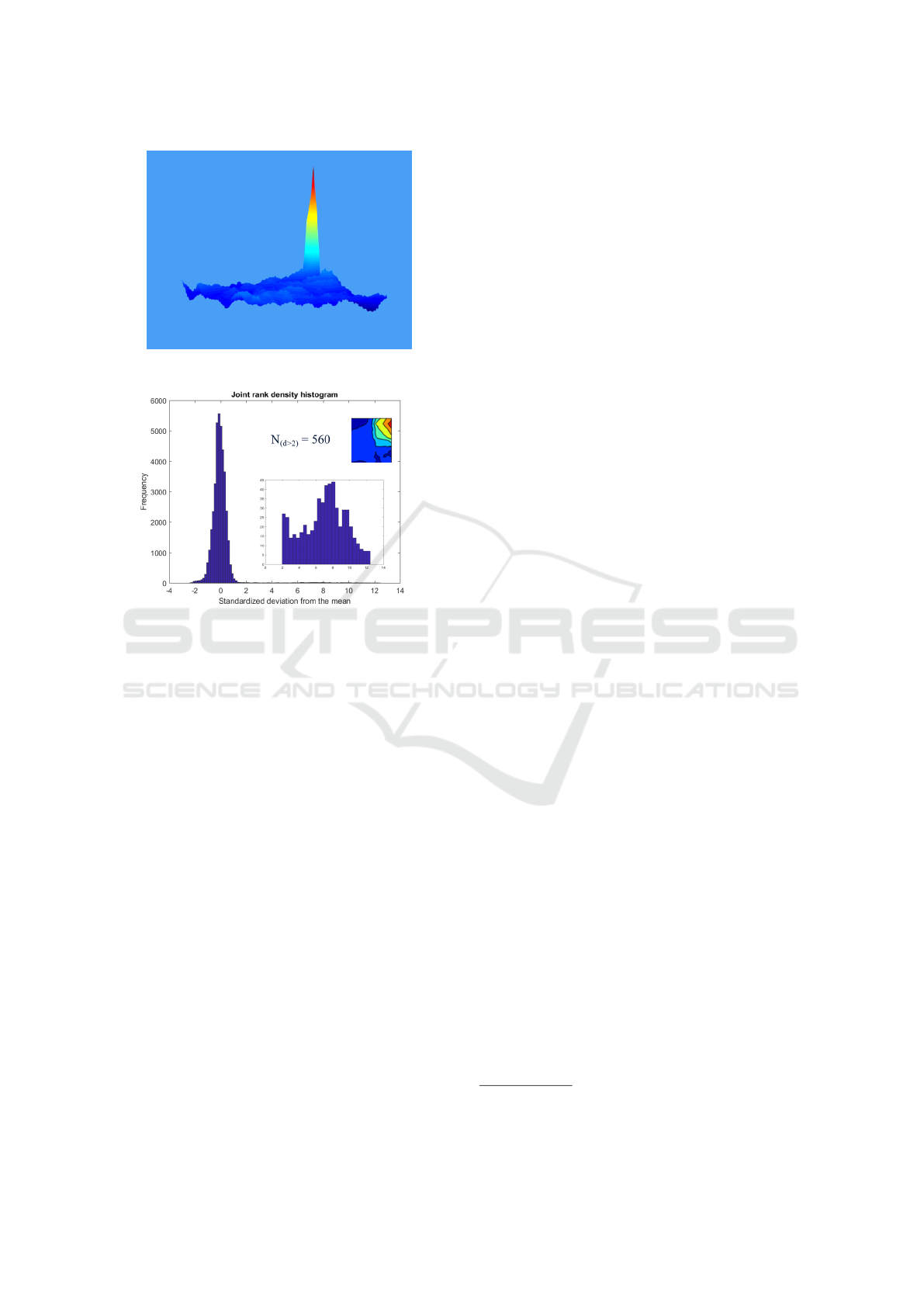
Figure 10: Joint rank density 3-d.
Figure 11: Joint rank density histogram.
5 PERFORMANCE
Computing the SVD in each ordination is the most
time consuming element of the method and the most
critical obstacle to scaling for Industrial Big Data. In
a series of experiments we tested three different im-
plementations and measured the performance as ex-
ecution time iterating over an increasing number of
dataset rows. Matlab code for the three implemen-
tations is shown in Sections 5.1 to 5.3. Tests were
run using Matlab(R) Version 9.3.0.713579 (R2017b)
on an ordinary desktop computer with an Intel(R)
Core(TM) i7-4790 CPU @ 3.60GHz, 3601 Mhz, and
Installed Physical Memory (RAM) of 16.0 GB.
Results of the performance tests are shown in Fig-
ure 12. The ‘naive’ but straightforward implemen-
tation following the mathematical notation and using
full diagonal matrices performs poorly, and is unus-
able for Big Data unless modified. Both the sparse
and vectorized implementations perform adequately
(vectorized being slightly better). These tests demon-
strate that the method does scale adequately to Indus-
trial Big Data, however further improvements by opti-
mizing the core SVD implementations should still be
investigated.
5.1 Full Diagonal Matrices
Notation follows Greenacre (2013, pp. 110–111).
Dr = diag(r);
% full diagonal
Dc = diag(c);
% full diagonal
T = Drˆ(-1/2)*(P-r*c’)*Dcˆ(-1/2);
% standardized residuals (Equation 4)
[U,S,V] = svd(T,0);
% singular value decomposition (economy size)
F = Drˆ(-1/2)*U*S;
% principal coordinates of the rows
5.2 Sparse Diagonal Matrices
Modifications to overcome the prohibitive size of the
diagonal matrices
2
.
rsq = 1./sqrt(r);
% inverse square root
nr = 1:numel(r);
% indices for sparse
rsp = sparse(nr,nr,rsq);
% sparse diagonal
csq = 1./sqrt(c);
% inverse square root
nc = 1:numel(c);
% indices for sparse
csp = sparse(nc,nc,csq);
% sparse diagonal
T = rsp*(P-r*c’)*csp;
% standardized residuals (Equation 4)
[U,S,V] = svd(T,0);
% singular value decomposition (economy size)
F = rsp*U*S;
% principal coordinates of the rows
5.3 Vectorised Loops
Modifications to improve speed as well as overcom-
ing the prohibitive size of the diagonal matrices
3
.
rsq = 1./sqrt(r);
% inverse square root
csq = 1./sqrt(c);
% inverse square root
T = P-r*c’;
for i = 1:size(T,2)
T(:,i)=(T(:,i).*rsq(:))*csq(i);
% standardized residuals (Equation 4)
end
[U,S,V] = svd(T,0);
% singular value decomposition (economy size)
F = U; s = diag(S);
for i = 1:size(F,2)
F(:,i) = (F(:,i).*rsq(:))*s(i);
% principal coordinates of the rows
end
2
Suggested by Odwa Sihlobo, Prescient SA.
3
Suggested by Stef Salvini, University of Oxford.
Anomaly Detection for Industrial Big Data
291

Figure 12: Performance evaluation plot.
6 CONCLUSIONS
In this paper we introduce a method for anomaly de-
tection to operate at industrial scale. The method has
relevance for system condition monitoring, prognos-
tic health maintenance, or any application where iden-
tification of anomalous cases is advantageous. The
method aligns with the general trend in Industrial Big
Data applications towards data-driven discovery as an
alternative to process models which are unlikely to
operate at the scales required. The method can ac-
commodate high-dimensional data of heterogeneous
data types in a simple and scalable computational
framework of well-known and well understood ordi-
nation techniques (e.g. SVD, PCA, CA; Greenacre
(2010, 2013)), and spatial density analysis (e.g. Bad-
deley et al. (2015)).
The method has a well defined workflow (refer to
Figure 1 and to the detailed descriptions in Section
2) and we demonstrate the method using data from
electricity smart meters (refer to Sections 3 and 4 and
to Figures 2 to 11). For the demonstration dataset
we find a small number (N
(d>2)
= 560; <0.5%) of
cases with overwhelming indication that they origi-
nate from a mechanism that is different from the one
that produced the bulk of cases in the dataset.
This process discovers those cases most resistant
to ordination, and therefore least conforming to the
mechanism that generated the rest of the data, i.e. the
anomalies.
In future work we plan to explore:
(i) the statistical properties of the method, specif-
ically the departure from the mean background
process as measured by the joint rank density of
cases,
(ii) further optimization for operating at scale of
the well-known algorithms for singular value
decomposition (SVD) as applied in the special
case of the method, and
(iii) the properties of the method when applying
bootstrapped resampling as described in Sec-
tion 2.1.2.
ACKNOWLEDGEMENTS
Funding in support of this work was given by Inno-
vate UK, Grant Reference Number 102510. Useful
comments on the manuscript were provided by Laura
Shemilt and Glen Moutrie.
DISCLOSURE
The technical method described in this paper is pro-
tected by British patent applications 1713703.5 and
1713896.7, submitted on 25 and 30 August 2017.
REFERENCES
Aggarwal, C. C. (2013). Outlier Analysis. Springer, Berlin.
Akoglu, L., Tong, H., and Koutra, D. (2015). Graph-based
anomaly detection and description: a survey. Data
Mining and Knowledge Discovery, 29:626–688.
Baddeley, A., Rubak, E., and Turner, R. (2015). Spatial
Point Patterns: Methodology and Applications with R.
Chapman and Hall/CRC Press.
Barnett, V. and Lewis, T. (1994). Outliers in Statistical
Data. Wiley, New York, 3rd. edition.
Campos, G. O., Zimek, A., Sander, J., Campello, R. J.
G. B., Micenkov, B., Schubert, E., Assent, I., and
Houle, M. E. (2016). On the evaluation of unsuper-
vised outlier detection: measures, datasets, and an em-
pirical study. Data Mining and Knowledge Discovery,
30:891–927.
Chandola, V., Banerjee, A., and Kumar, V. (2009).
Anomaly detection: a survey. ACM Computing Sur-
veys, 41:1–58.
de Mauro, A., Greco, M., and Grimaldi, M. (2015). What
is big data? A consensual definition and a review
of key research topics. AIP Conference Proceedings,
1644(1):97–104.
Efron, B. and Tibshirani, R. J. (1994). An Introduction to
the Bootstrap. Chapman and Hall/CRC, New York.
Greenacre, M. (2010). Biplots in Practice. BBVA Founda-
tion, Madrid.
Greenacre, M. (2013). Contribution biplots. Journal of
Computational and Graphical Statistics, 22:107–122.
Hawkins, D. (1980). Identification of Outliers. Chapman
and Hall, London.
DATA 2018 - 7th International Conference on Data Science, Technology and Applications
292

Jackson, D. A. (1993). Stopping rules in principal compo-
nents analysis: A comparison of heuristical and statis-
tical approaches. Ecology, 74:2204–2214.
Knorr, E. M. and Ng, R. T. (1997). A unified notion of
outliers: properties and computation. Proceedings of
the 3rd ACM international conference on knowledge
discovery and data mining (KDD), Newport Beach.
Nunnaly, J. C. and Bernstein, I. H. (1994). Psychometric
theory. McGraw-Hill, New York, 3rd edition.
Peres-Neto, P. R., Jackson, D. A., and Somers, K. M.
(2005). How many principal components? Stopping
rules for determining the number of non-trivial axes
revisited. Computational Statistics and Data Analy-
sis, 49:974–997.
Schubert, E., Zimek, A., and Kriegel, H. P. (2014). Local
outlier detection reconsidered: a generalized view on
locality with applications to spatial, video, and net-
work outlier detection. Data Mining and Knowledge
Discovery, 28:190–237.
Anomaly Detection for Industrial Big Data
293
