
Multiclass Tissue Classification of Whole-Slide Histological Images using
Convolutional Neural Networks
Rune Wetteland
1
, Kjersti Engan
1
, Trygve Eftestøl
1
, Vebjørn Kvikstad
2
and Emilius A. M. Janssen
2,3
1
Department of Electrical Engineering and Computer Science, University of Stavanger, Norway
2
Department of Pathology, Stavanger University Hospital, Norway
3
Department of Mathematics and Natural Sciences, University of Stavanger, Norway
Keywords:
Histological Whole-Slide Images, Autoencoder, Deep Learning, Semi-supervised Learning, ROI Extraction.
Abstract:
Globally there has been an enormous increase in bladder cancer incidents the past decades. Correct prognosis
of recurrence and progression is essential to avoid under- or over-treatment of the patient, as well as unnec-
essary suffering and cost. To diagnose the cancer grade and stage, pathologists study the histological images.
However, this is a time-consuming process and reproducibility among pathologists is low. A first stage for an
automated diagnosis system can be to identify the diagnostical relevant areas in the histological whole-slide
images (WSI), segmenting cell tissue from damaged areas, blood, background, etc. In this work, a method
for automatic classification of urothelial carcinoma into six different classes is proposed. The method is based
on convolutional neural networks (CNN), firstly trained unsupervised using unlabelled images by utilising an
autoencoder (AE). A smaller set of labelled images are used to train the final fully-connected layers from the
low dimensional latent vector of the AE, providing an output as a probability score for each of the six classes,
suitable for automatically defining regions of interests in WSI. For evaluation, each tile is classified as the class
with the highest probability score. The model achieved an average F1-score of 93.4% over all six classes.
1 INTRODUCTION
Globally, bladder cancer resulted in 123,400 deaths in
1990, and in 2010 this number was 170,700 which is
an increase of 38,3% taking population growth into
consideration (Lozano et al., 2012). The majority
of bladder cancer incidents are urothelial carcinoma
with a representation as high as 90% in some regions
(Eble et al., 2004). For patients diagnosed with blad-
der cancer, 50-70% will experience one or more re-
currences, and 10-30% will have disease progression
to a higher stage (Mangrud, 2014). Patient treatment,
follow-up and calculating the risk of recurrence and
disease progression depend primarily on the histolog-
ical grade and stage of cancer. Correct prognosis of
recurrence and progression is essential to avoid under-
or over-treatment of the patient, as well as unneces-
sary suffering and cost.
With the introduction of digital pathology, some
computer-aided tools to assist pathologists have been
introduced, but still the assessment of histopatholog-
ical images to diagnose, grade and stage cancer is
mainly done manually. This is a time-consuming
process and reproducibility among pathologists is in
some cases low, for example within the prognostic
classification of urinary bladder cancer. Automatic
extraction of the relevant areas in large whole-slide
images (WSI) would be an important first step where
the results could be used in automated diagnostic and
prognostic classification tools.
During the biopsy, parts of the tissue get both
physical- and heating-damage, and thus can not be
used as relevant diagnostic information. The WSI
also contains stroma- and muscle-tissue as well as ar-
eas of blood. In this paper we consider the task of
automatic classification of tiles in WSI into the six
different classes; urothelium, stroma, damaged tissue,
muscle, blood and background. Examples from each
class are shown in Figure 1. The system uses the au-
tomatic classification tool to produce heat maps from
the model’s output. Such heat maps can provide use-
ful information to help the pathologist to focus on the
diagnostic important part of the large WSI during vi-
sual inspection. In addition, the heat maps are also
suitable as input for automatic region of interest (ROI)
extraction of relevant areas in the WSI, which can fur-
ther be used in automated diagnostic and prognostic
classification tools.
320
Wetteland, R., Engan, K., Eftestøl, T., Kvikstad, V. and Janssen, E.
Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks.
DOI: 10.5220/0007253603200327
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 320-327
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 1: Example tiles from each class. A) Urothelium, B) Stroma, C) Damaged tissue, D) Muscle tissue, E) Blood, and F)
Background.
1.1 Previous Work
In recent literature, some methods for automatic tis-
sue classification have been suggested. However,
most previous works have focused on classifying only
two classes, a binary problem set to differentiate be-
tween cancer-patches and non-cancer patches.
Recent literature shows good results for binary tis-
sue classification using convolutional neural networks
(CNN). Wang et al. (2016) won both competitions of
the Camelyon16 grand challenge for automated de-
tection of metastatic breast cancer in WSI. As part
of their model, GoogLeNet was utilised to do patch
classification. The model was trained to discriminate
between positive and negative patches and achieved
an accuracy of 98.4%.
Some attempts of multiclass tissue classification
can be found in recent years. Araujo et al. classified
patches of breast cancer into four classes using con-
volutional neural networks (Ara
´
ujo et al., 2017). The
best patch-wise accuracy for four classes was 66.7%.
When the task was simplified as a two-classes prob-
lem, non-carcinoma vs carcinoma, the accuracy was
improved to 77.6%. The work of Kather et al. (2016)
uses a combination of several hand-crafted feature
methods to classify different types of tissue in col-
orectal cancer, performing tests on both a two-class
and eight-class problem. They achieved the best re-
sult on the two-class problem with a tumour-stroma
separation accuracy of 98.6%, while the multiclass
problem achieved an accuracy of 87.4%.
To the author’s knowledge, there are no published
results on multiclass classification on WSI of bladder
cancer.
Some few and recent work on ROI detection can
be found. ROI detection has been done by multi-scale
real-time coarse-to-fine topology preserving segmen-
tation (CTFTPS) by utilising superpixel clustering
technique (Li and Huang, 2015; Yao et al., 2015).
A RAPID (Regular and Adaptive Prediction-Induced
Detection) segmentation method for ROI detection in
large WSI is presented by Sulimowicz and Ahmad
(2017) while using the multi-scale CTFTPS technique
as a baseline. An SVM was utilised to classify the
detected regions as ROI vs non-ROI. For this task,
the classifier achieved an F1-score of 89.8% for the
RAPID method, and 91.2% for the optimised multi-
scale CTFTPS method.
Deep CNN has shown to provide state of the
art results in many computer vision tasks in recent
years (LeCun et al., 2015) and has also found its way
into medical image assessment tasks. In this work,
a method for automatic classification of WSI from
urothelial carcinoma into six different classes is pro-
posed. The method is based on CNN, firstly trained
unsupervised, using large unlabelled image sets by
utilising an autoencoder (AE). A set of labelled im-
ages are used to train the final fully-connected layers
from the low dimensional latent vector of the AE, pro-
viding an output as a probability score for each of the
Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks
321

nostra, per inceptos himenaeos. Proin rutrum risus id sodales laoreet. Aenean tincidunt porta est. Ut suscipit, ligula tincidunt lacinia interdum, urna sapien rutrum orci, vitae consequat ex
elit at ipsum. Cras et arcu at justo ultrices tincidunt. Sed maximus libero at neque lobortis ultricies. Suspendisse quis quam nibh. Quisque rhoncus fringilla facilisis.
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Donec diam leo, consectetur ac feugiat eget, laoreet id eros. Phasellus lectus mi, scelerisque ac finibus a, dictum id arcu. Nunc
vehicula ante erat, id faucibus sapien placerat id. Vivamus ultricies diam non magna iaculis gravida. Cras lacinia scelerisque lacus, non consequat nisl hendrerit nec. Aliquam viverra, nisl
ut pretium egestas, metus dolor feugiat purus, in malesuada sem quam id neque. Donec eleifend vestibulum enim at auctor. Class aptent taciti sociosqu ad litora torquent per conubia
nostra, per inceptos himenaeos. Proin rutrum risus id sodales laoreet. Aenean tincidunt porta est. Ut suscipit, ligula tincidunt lacinia interdum, urna sapien rutrum orci, vitae consequat ex
elit at ipsum. Cras et arcu at justo ultrices tincidunt. Sed maximus libero at neque lobortis ultricies. Suspendisse quis quam nibh. Quisque rhoncus fringilla facilisis.
Decoder
Classifier
128x128x8
64x64x8
Reconstructed tile
(128x128x3)
32x32x8
4096
2048
8192
Classifier
6) Background
1 %6
5) Blood
4) Muscle tissue
3)Damagedtissue
2) Stroma
1) Urothelium
1 %
5
2 %4
6 %3
21 %
2
69 %1
Probability
output
256
6
Encoder
128x128x8
64x64x8
Input tile
(128x128x3)
4096
2048
32x32x8
Latent
vector
1024
Unlabelled dataset
Max PoolingConvolution Dropout Fully-connected Deconvolution Unpooling
Whole-Slide Images
Labelled dataset
512
Figure 2: Overview of the CNN-model. First, the unlabelled dataset is used to train the encoder-decoder model. Then the
labelled dataset is used to train the encoder-classifier model. Finally, the trained encoder-classifier model are used to classify
new WSI into probability maps. These probability maps are further postprocessed to produce the heat maps.
six classes, suitable for automatically defining ROI in
WSI. A visualisation of the system is depicted in Fig-
ure 2.
The novelty of the work lies both in the specific
application of urinary bladder WSI and in the method
development, more specifically in a combination of
using CNN, learned in a semi-supervised way, for the
application of automatic region of interest extraction
in WSI by multiclass tissue classification, tested on
urinary bladder cancer.
2 DATA MATERIAL
The data material used in this paper consists of
histopathological images from patients with primary
bladder cancer, collected in the period 2002-2011 at
the University Hospital of Stavanger in Norway. The
biopsies are formalin fixed and paraffin embedded, 4
µm slides are cut and stained with Hematoxylin Eosin
Saffron (HES). All slides are diagnosed and graded
according to WHO73 and WHO04, cancer stage (Tis,
Ta or T1) and follow-up data on recurrence and dis-
ease progression are recorded.
The slides are then scanned using a Leica SCN400
histological slide scanner to produce a digital histo-
logical image. The images are in Leicas data format
called SCN and to be able to process these images
the Vips library (Martinez and Cupitt, 2005) has been
used, which is specially designed for image
processing of large images.
3 PROPOSED METHOD
An overview of the proposed method can be seen in
Figure 2. The different parts will be explained in this
section.
3.1 Preprocessing
Each WSI is sliced into smaller non-overlapping tiles
of size 128x128 pixels, extracted at 400x magnifica-
tion level. The background takes up as much as 70-
80% of the WSI and is detected and discarded auto-
matically by computing the histogram of the tile and
setting a fixed threshold value. This removes tiles
consisting of grey background, however, if the back-
ground tile contains small parts of debris, tissue or
similar it is not discarded. Examples of tiles belong-
ing to this class are illustrated in Figure 1-F.
The histological images are split into three
datasets. First, an unlabelled dataset is created in
the manner explained above where the extracted tiles
have no label associated with it. In total 48 WSI all
from different patients were preprocessed resulting in
7,130,527 unlabelled tiles after the pure background
tiles are excluded. This set, called train-ae, is utilised
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
322

as training data for the AE-model.
Secondly, a labelled training dataset is created. A
pathologist has manually annotated carefully selected
regions in the WSI. The tiles in the regions are pre-
processed by evaluating the histogram to be sure not
to include background or boundaries and given a la-
bel corresponding to its class. The number of patients
and tiles produced are listed as train-set1 in Table 1.
Lastly, a labelled test set is created to assess the
performance of the classifier. The set is created in
the same manner as the labelled training set, but
on separate WSI which has not been used in either
the unlabelled or labelled datasets to avoid cross-
contamination between training and test data. The
dataset is listed as test-set in Table 1.
The texture of urothelium tissue will change for
the different cancer grades, and thus it is vital to in-
clude a wide variety of samples for this class. The
other five classes, however, will not change as a func-
tion of cancer grade and may include fewer samples.
Another issue is that the occurrence of some classes is
more sparse in the WSI, making it difficult to extract a
large amount of it. A disadvantage of these two issues
is a significant deviation in the number of samples in
two of the classes, stroma and muscle tissue, as seen
in train-set1 in Table 1.
To compensate for the class-imbalance in train-
set1, data augmentation techniques have been utilised.
Tiles in the muscle and stroma class are extracted with
50% overlap, to produce more data from the same re-
gions. These extracted tiles are further augmented by
randomly flipping and rotating them to create new
data. These techniques result in a more balanced
dataset, which is listed in Table 1 as train-set2. This
dataset is used to train the classifier in the presented
experiments. The augmentation techniques were not
performed on the test-set, resulting in an unbalanced
test set. In this case, accuracy as a performance met-
ric could be misleading. Instead, precision, recall and
F1-score are used to evaluate the performance.
Table 1: The resulting labelled datasets after preprocessing.
Results show the total number of tiles extracted for each
class, and the number of WSI used are shown in parenthese.
Train-set1 Train-set2 Test-set
Urothelium 25,635 (25) 25,635 (25) 3,612 (3)
Stroma 4,329 (4) 25,974 (4) 505 (1)
Damaged 30,714 (8) 30,714 (8) 2,679 (1)
Muscle 2,002 (3) 23,949 (3) 475 (1)
Blood 19,071 (4) 19,071 (4) 692 (1)
Background 20,000 (2) 20,000 (2) 500 (1)
3.2 CNN-Model
The system consists of an autoencoder model which
is trained on the unlabelled dataset train-ae. The au-
toencoder consists of two main parts; the encoder
and the decoder. The encoder will transform the in-
put tile into a latent vector of much lower dimension.
A small latent space is chosen which will force the
network to extract the essential features of the image
and preserve these in the vector. The decoder will
use the features stored in the latent vector and recon-
struct the input. During training, the network com-
pares the reduced mean of the squared difference be-
tween the input image and reconstructed output im-
age as given by the loss function
∑
(input − out put)
2
.
The AE function is described in details in (Baldi,
2012). The encoder consists of two convolutional-,
two max-pooling- and four dropout-layers, as well as
three fully-connected layers as seen in Figure 2. The
decoder consists of the same layers, but in reverse or-
der and uses unpooling and deconvolutional layers in-
stead.
After training, the encoder has learned to extract
the features of the input tile, which are now stored
in the latent vector. To do classification, the decoder
part is discarded and exchanged with a classifier. The
classifier consists of three fully-connected layers con-
nected to the output of the encoder. This encoder-
classifier model constitutes the proposed CNN-model
and is trained on the labelled training dataset train-
set2 and evaluated on the test-set.
For initialisation of the system, the bias is set to
zero, and the weights are taken from a truncated nor-
mal distribution. The convolutional layers use a fil-
ter kernel of 3x3 and a stride of 1, whereas the max-
pooling layers use a filter kernel of 2x2 with a stride
of 2. The number of feature maps is used to control
the size of the latent vector space and is experimented
on as described in section 4. The parameters of the
network are optimised using the Adam optimiser with
a mini-batch of size 128. For the activation function
between layers, the Rectified linear unit (ReLU) acti-
vation function is used. For the last layer, the Softmax
activation function is utilised. This will output a true
probability distribution, meaning each output lays in
the interval 0 to 1 and all outputs combined sums up
to one. Dropout is a technique where randomly se-
lected nodes are set to zero during training to provide
regularisation to the network. The portion of nodes
set to zero is specified by the dropout rate as a per-
centage. During evaluation of the network, dropout is
disabled.
The histological images are in Leicas data format
called SCN and to be able to process these images
Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks
323

the Vips library (Martinez and Cupitt, 2005) has been
used. This is a library specially designed for image
processing of large images. The model is written in
Python 3.5 using the Tensorflow 1.7 machine learn-
ing library (Abadi et al., 2016). For evaluation of
the model, the Scikit-learn metric package (Pedregosa
et al., 2011) is used which computes precision, recall
and F1-score of each class in addition to an average
total score.
The model is used to predict the class of each tile
in a WSI. The probability for each class provided by
the model can be rearranged as probability maps, one
for each class, and will visualise the location in the
histological image where each class is present. An
overview of this process is presented in Figure 2.
4 EXPERIMENTS AND RESULTS
Two experiments were conducted, the first to find the
best combination of architecture and hyperparameters
and the second to verify its performance and use the
final model on WSI.
4.1 Experiment 1: Architecture and
Hyperparameters
To find a suitable architecture and appropriate hyper-
parameters, a large grid search was conducted. To
reduce both computational time and search space, a
preliminary search was set up with some limitations.
A reduced version of the train-ae dataset was used
to decrease the processing time, and each model was
only trained for 50 epochs.
The encoder-decoder model was tested with two
different sizes of the latent vector, which was altered
by changing the number of feature maps in the con-
volutional layers. Latent vectors of size 512 and 1024
were tested. A learning rate of 10
-3
and 10
-4
was
tested as well as dropout rates of 0%, 10% and 20%.
Each of these combinations was tested on network
configuration consisting of two, four and six convo-
lutional layers in the autoencoder.
In the encoder-classifier model, the classifier con-
sists of three dense layers. The first layer after the en-
coder was tested with 256, 512 and 1024 neurons, and
the second layer with 128, 256 and 512 neurons. The
number of neurons in the output layer is bounded to
the number of classes. This results in 9 different con-
figurations for the classifier layers. Each of these con-
figurations was tested with a learning rate of 10
-3
, 10
-4
and 10
-5
. There are no dropout layers in the classifier
itself, but changing the dropout rate will affect how
the encoder codes the input tile into the latent vec-
tor. The encoder-classifier was therefore also tested
with the same dropout rates as above. The model was
tested both with and without freezing the pre-trained
encoder-layers to see how it affected the result.
The prediction accuracy on the test-set was used
to compare the performance of the different hyperpa-
rameter combinations. Hyperparameters that showed
poor performance on several models were excluded to
narrow down the search space.
The experiments showed an overall best result us-
ing an encoder-decoder structure with two convolu-
tional layers with a latent vector of 1024 neurons
trained with 10
-4
learning rate and 10% dropout rate.
The results further showed best performance while
not freezing the encoder part of the encoder-classifier
model. A classifier with 256 neurons in the first layer
and 512 in the second layer was favourable, trained
using a learning rate of 10
-5
and 10% dropout rate.
These hyperparameters and settings will be used as
the resulting model of this experiment. The model is
depicted in Figure 2.
4.2 Experiment 2: Training, Testing
and using the Resulting Model
The resulting architecture after the first experiment
was trained once more, this time on the full dataset.
First, the autoencoder was trained on the unlabelled
dataset train-ae for 100 epochs, then the encoder-
classifier was fine-tuned on the augmented labelled
dataset train-set2 for another 600 epochs. Since ex-
periment 1 showed best results when the encoder was
not frozen during fine-tuning, both the encoder and
classifier was trained during this step. Evaluation us-
ing the Scikit-learn metric package on the test-set was
performed every 5th epoch. The model achieved the
best result after 540 epochs of training with an aver-
age F1-score of 93.4% over all six classes. The pre-
cision, recall and F1-score of each class is shown in
Table 2.
Table 2: Detailed classification results from the model
trained using 10% dropout rate.
Class Precision Recall F1-Score
Urothelium 0.924 0.952 0.938
Stroma 0.897 0.929 0.913
Damaged 0.925 0.927 0.926
Muscle 0.980 0.714 0.826
Blood 0.996 0.991 0.994
Background 0.990 0.988 0.989
Average total 0.936 0.935 0.934
The overall results in Table 2 are good. However,
there are some observations.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
324

In train-set2, which is used to train the classifier,
the classes of blood and background have the fewest
number of samples. However, these are the classes
which perform best. This is probably because these
classes have the least within-class variance, e.g. most
of the tiles have a similar visual appearance.
Urothelium and damaged tissue both perform
well, even though these classes have a substantial vi-
sual variance in the form of colour and texture in the
tiles. The dataset for these classes contains the most
number of patients (25 and 8 patients, respectively),
and therefore contains the most diverse samples in the
dataset, contributing to the good results.
The precision of stroma and recall of muscle is
not performing as good as the rest. The dataset for
these classes contains few patients and are also the
two classes which needed augmentation due to small
amounts of available data. The low recall of muscle
tissue indicates that a large proportion of the muscle
tiles are misclassified as other classes, most proba-
bly urothelium, stroma and damaged tissue (due to the
high precision of blood and background, these are not
likely to include many misclassified tiles). It is im-
portant to note that the muscle class achieves a very
good precision score, and stroma has an acceptable
good recall score.
4.3 Heat Maps
The resulting model was utilised to classify entire
whole-slide images. Each tile in the WSI was classi-
fied and the percentage for each class recorded. These
were then combined to create the probability maps.
These maps were then post-processed in MATLAB by
applying a Gaussian filter kernel with a standard devi-
ation of σ = 0.6 to smooth the images. After filtering,
a thresholding operation was performed on the image
with a limit of 0.8, setting all predictions below this
threshold to zero. This ensures that only predictions
of 0.8 or higher are visible in the final heat maps.
Figure 3 shows three example WSI with their
corresponding heat maps. By visual inspection per-
formed by pathologists, this is considered to look very
promising. However, a quantitative measure for the
WSI ROI extraction is lacking since we do not have
complete WSI manually labelled into the six classes
at the current time.
5 CONCLUSION
This paper proposes a method for automatic classi-
fication of tile-segments of histopathological WSI of
urinary bladder cancer into six different classes us-
ing a CNN-based model. An encoder-decoder struc-
ture is trained on a large set of unlabelled data. After
training, the encoder part of the autoencoder acts as a
feature extractor making low dimensional latent vec-
tors. An encoder-classifier structure is then fine-tuned
on a set of labelled tiles. The finished model is able
to classify input tiles from the WSI into the classes
urothelium, stroma, damaged tissue, muscle, blood
and background. The best model achieved an average
F1-score of 93.4% over all the six classes, an overall
good result. However, future work will include an ef-
fort to improve the classifier. Other methods such as
a multiscale approach are considered.
The model is further used to classify entire WSI
to produce heat maps, which visualises each of the
classes and their location in the image. These maps
can provide useful information to the pathologist dur-
ing visual inspection. Future work consists of using
the above model as an ROI extractor of relevant tissue
in the WSI to make a dataset suitable as training data
for a diagnostic and prognostic classification model.
REFERENCES
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean,
J., Devin, M., Ghemawat, S., Irving, G., Isard, M.,
et al. (2016). Tensorflow: a system for large-scale
machine learning. In OSDI, volume 16, pages 265–
283.
Ara
´
ujo, T., Aresta, G., Castro, E., Rouco, J., Aguiar,
P., Eloy, C., Pol
´
onia, A., and Campilho, A. (2017).
Classification of breast cancer histology images us-
ing convolutional neural networks. PloS one,
12(6):e0177544.
Baldi, P. (2012). Autoencoders, unsupervised learning, and
deep architectures. In Proceedings of ICML workshop
on unsupervised and transfer learning, pages 37–49.
Eble, J. N., Sauter, G., Epstein, J. I., and Sesterhenn, I. A.
(2004). World Health Organization Classification of
Tumours. Pathology and Genetics of Tumours of the
Urinary System and Male Genital Organs. IARC
Press: Lyon.
Kather, J. N., Weis, C.-A., Bianconi, F., Melchers, S. M.,
Schad, L. R., Gaiser, T., Marx, A., and Z
¨
ollner, F. G.
(2016). Multi-class texture analysis in colorectal can-
cer histology. Scientific reports, 6:27988.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. nature, 521(7553):436.
Li, R. and Huang, J. (2015). Fast regions-of-interest de-
tection in whole slide histopathology images. In In-
ternational Workshop on Patch-based Techniques in
Medical Imaging, pages 120–127. Springer.
Lozano, R., Naghavi, M., Foreman, K., and Lim, S. (2012).
Global and regional mortality from 235 causes of
Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks
325
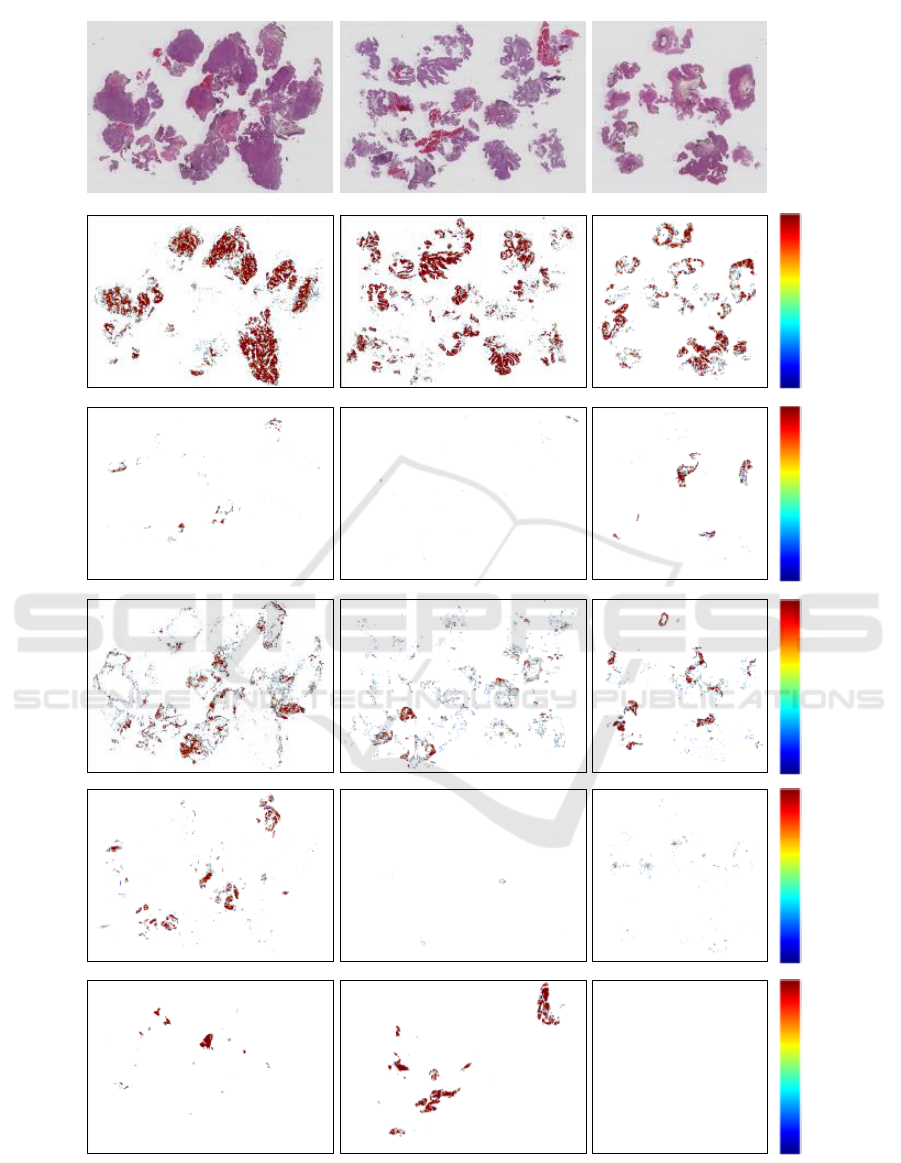
Original image
Stroma
Urothelium
Damaged
Blood
Muscle
0.8
1.0
0.9
0.8
1.0
0.9
0.8
1.0
0.9
0.8
1.0
0.9
0.8
1.0
0.9
Figure 3: The original WSI together with the corresponding heat maps. The scale in the rightmost column shows the con-
fidence level given by the model. The background heat maps are performing very good, but has been omitted from the heat
map visualisation since it is just removing the borders between background and tissue. The heat maps have been smoothed
with a Gaussian filter and thresholded to only contain predictions of 0.8 and higher.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
326

death for 20 age groups in 1990 and 2010: a system-
atic analysis for the Global Burden of Disease Study
2010. The Lancet, 380(9859):2095–2128.
Mangrud, O. (2014). Identification of patients with high
and low risk of progresson of urothelial carcinoma of
the urinary bladder stage Ta and T1. PhD thesis, Ph.
D. dissertation, University of Bergen.
Martinez, K. and Cupitt, J. (2005). Vips-a highly tuned im-
age processing software architecture. In Image Pro-
cessing, 2005. ICIP 2005. IEEE International Confer-
ence on, volume 2, pages II–574. IEEE.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., et al. (2011). Scikit-
learn: Machine learning in python. Journal of ma-
chine learning research, 12(Oct):2825–2830.
Sulimowicz, L. and Ahmad, I. (2017). “rapid” regions-
of-interest detection in big histopathological images.
In Multimedia and Expo (ICME), 2017 IEEE Interna-
tional Conference on, pages 595–600. IEEE.
Wang, D., Khosla, A., Gargeya, R., Irshad, H., and Beck,
A. H. (2016). Deep learning for identifying metastatic
breast cancer. arXiv preprint arXiv:1606.05718.
Yao, J., Boben, M., Fidler, S., and Urtasun, R. (2015).
Real-time coarse-to-fine topologically preserving seg-
mentation. In Proceedings of the IEEE Conference
on Computer Vision and Pattern Recognition, pages
2947–2955.
Multiclass Tissue Classification of Whole-Slide Histological Images using Convolutional Neural Networks
327
