
Eliminating Noise in the Matrix Profile
Dieter De Paepe, Olivier Janssens and Sofie Van Hoecke
Ghent University – imec – IDLab, Department of Electronics and Information Systems, Ghent, Belgium
Keywords:
Matrix Profile, Noise, Time Series, Anomaly Detection, Time Series Segmentation.
Abstract:
As companies are increasingly measuring their products and services, the amount of time series data is rising
and techniques to extract usable information are needed. One recently developed data mining technique for
time series is the Matrix Profile. It consists of the smallest z-normalized Euclidean distance of each subse-
quence of a time series to all other subsequences of another series. It has been used for motif and discord
discovery, for segmentation and as building block for other techniques. One side effect of the z-normalization
used is that small fluctuations on flat signals are upscaled. This can lead to high and unintuitive distances for
very similar subsequences from noisy data. We determined an analytic method to estimate and remove the
effects of this noise, adding only a single, intuitive parameter to the calculation of the Matrix Profile. This
paper explains our method and demonstrates it by performing discord discovery on the Numenta Anomaly
Benchmark and by segmenting the PAMAP2 activity dataset. We find that our technique results in a more
intuitive Matrix Profile and provides improved results in both usecases for series containing many flat, noisy
subsequences. Since our technique is an extension of the Matrix Profile, it can be applied to any of the various
tasks that could be solved by it, improving results where data contains flat and noisy sequences.
1 INTRODUCTION
The amount of data available as time series is rapidly
increasing. This increase can be explained by the
lowering cost of sensors and storage, but also due to
the rising interest of companies to gain new insights
about their products or services. Areas of interest in-
clude pattern discovery (Papadimitriou and Faloutsos,
2005), anomaly detection (Wang et al., 2016) and user
load prediction (Vandewiele et al., 2017).
The Matrix Profile is a recently developed data
mining technique for time series data that is calcu-
lated using two time series and a single intuitive pa-
rameter: the subsequence length m. It is a one-
dimensional series where each data point at a given
index represents the Euclidean distance between the
z-normalized (zero mean and unit variance) subse-
quence starting at that index in the first time series
and the best matching (lowest distance) z-normalized
subsequence in the second time series. Both inputs
can be the same, meaning matches are searched for in
the same time series, in this case a trivial match buffer
prevents the subsequence matching itself or nearby
neighbours. The Matrix Profile Index, which is calcu-
lated alongside the Matrix Profile, contains the index
of the best match for each subsequence.
The Matrix Profile can be used for motif discovery
(finding the best matching subsequence in a series),
discord discovery (finding the subsequence with the
largest distance to its nearest match) or as a building
block for other techniques such as segmentation, vi-
sualizing time series using Multidimensional Scaling
(MDS) (Yeh et al., 2017b) or finding gradually chang-
ing patterns in time series (Time Series Chains) (Zhu
et al., 2017). The Matrix Profile has been applied to
various topics, including medical signals (Yeh et al.,
2016), query logs (Zhu et al., 2017) and music (Yeh
et al., 2017a).
The original STAMP algorithm is able to calcu-
late the Matrix Profile in O(n
2
logn) time and O(n)
memory (Yeh et al., 2016). It uses the fact that the
z-normalized Euclidean distance between two vectors
can be calculated in constant time, given the dot prod-
uct of those two vectors, it calculates these dot prod-
ucts using the MASS algorithm (Mueen et al., 2015),
based on the Fast Fourier Transform. The later devel-
oped STOMP algorithm further improves this runtime
to O(n
2
) by reusing intermediate results.
A number of features/advantages of the Matrix
Profile are:
• The only required parameter is the subsequence
length m, which has a clear meaning for users;
• The runtime to calculate the Matrix Profile is in-
dependent of the chosen subsequence length m;
De Paepe, D., Janssens, O. and Van Hoecke, S.
Eliminating Noise in the Matrix Profile.
DOI: 10.5220/0007314100830093
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 83-93
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
83

Figure 1: Top: Three pairs of sequences with varying slopes
but exactly the same amount of noise. Bottom: The z-
normalized versions of the same sequences. We see how de-
pending on the slope, the Euclidean distance greatly varies
between the z-normalized sequences due to the amplifica-
tion of the noise. The z-normalized Euclidean distances are
(from left to right): 7.61, 3.55 and 1.81.
• Using the STAMP algorithm, a very accurate
guess of the Matrix Profile can be made in a frac-
tion of the time needed to calculate it;
• The algorithms to calculate the Matrix Profile are
well suited for parallelization, as has been demon-
strated in a GPU implementation (Zhu et al.,
2016);
• The Matrix Profile is not based on heuristics and
always provides an exact solution;
• No assumptions are made about the data, making
this technique domain agnostic.
Although z-normalization is important when com-
paring time series (Keogh and Kasetty, 2002), there is
one major downside: on a flat signal, any fluctuations
(noise) on the signal are enhanced, resulting in high
values in the Matrix Profile. We demonstrate this in
Figure 1: we created 3 pairs of sequences, each con-
sisting of two lines having a specific slope. Each pair
has the exact same amount of noise added to both
lines. Despite the original sequences being equally
distant, the z-normalized sequences have a lower dis-
tance on the sloped lines. Since the Matrix Profile
uses the z-normalized Euclidean distance, it is also
affected by this phenomenon.
Most use cases in literature relating to the Matrix
Profile focus on processing time series where flat se-
quences are rare or indicate periods of (uninterest-
ing) inactivity, these are not negatively affected by
this phenomenon. On the other hand, cases where flat
patterns are relevant will suffer from the influence of
noise. As a preliminary example, observe Figure 4.
Here, we see a discord that is not picked up by the
Matrix Profile due to the noise in the flat areas of the
signal. We discuss this example in detail in Section 4.
Generally, the effects of flat sequences might re-
sult in misleading or plain wrong insights. This is
problematic, since in many realistic cases a flat se-
quence can occur and might simply indicates a steady
regime of the underlying process. Some cases and
their respective periods of flat signals are: seismol-
ogy (periods of seismic inactivity), system monitor-
ing (similar metrics during a long running task - used
in Section 4) and movement monitoring (periods of
inactivity - used in Section 5). To improve the results
of the Matrix Profile for these kinds of cases, we in-
vestigated and analytically estimated the influence of
noise on the Matrix Profile, allowing us to negate it.
This paper is structured as follows: Section 2 lists
literature related to the Matrix Profile. In Section 3 we
explain how to eliminate the effects of noise. Section
4 demonstrates the effect of our technique on anomaly
detection for a simple synthetic example and using
data from the Numenta Anomaly Benchmark (NAB).
In Section 5 we apply our technique to time series
segmentation on passive activities using the PAMAP2
dataset (Reiss and Stricker, 2012). We conclude our
findings in Section 6.
2 RELATED WORK
In this section we focus on related work specifically
about the Matrix Profile.
The original Matrix Profile paper (Yeh et al.,
2016) introduces the Matrix Profile as a new method
for finding similarities in one-dimensional time series,
which can also serve for motif and discord discovery
as a side effect. The authors explain the STAMP and
STAMPI algorithms to calculate the Matrix Profile in
batch and incremental steps respectively, based on the
MASS algorithm (Mueen et al., 2015). A later paper
(Yeh et al., 2017a) extends the Matrix Profile to be
able to work with multi-dimensional data.
The STOMP algorithm, a faster version of
STAMP that reuses intermediary results in its calcu-
lation, is introduced in (Zhu et al., 2016). The same
paper describes how a GPU-based version of STOMP
(GPU-STOMP) was used to calculate the Matrix Pro-
file on a seismology dataset of 100 million datapoints
in only 12 days.
Yeh et al. describe how MDS, a data explo-
ration technique, does not work well when consid-
ering the subsequences in a time series (Yeh et al.,
2017b). Instead, it is useful to select representative
subsequences to use as input for MDS. When dealing
with discrete time series, the selection can be done
using the Minimum Description Length (MDL) and
Reduced Description Length (RDL), effectively se-
lecting subsequences that provide good compression.
The authors find that the Euclidean distance is a good
proxy for RDL in real-valued time series, and can be
used to select the salient subsequences for display in
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
84

MDS, meaning the Matrix Profile can be used for this
task.
Sometimes the findings of the Matrix Profile do
not coincide with the interests of the user, as they
might be interested in specific time frames, highly
variable patterns or in regions coinciding with activity
in another time series. Using a user-defined Annota-
tion Vector, introduced in (Dau and Keogh, 2017), a
user can modify a Matrix Profile, putting more focus
on the regions that he is interested in.
Time Series Chains are defined in (Zhu et al.,
2017) as slowly changing, repeated patterns in time
series. To find them, they define the left and right
Matrix Profile, containing the distance to the best ear-
lier and later subsequences. They are calculated using
a modified version of STOMP that has the same com-
plexity.
The Matrix Profile Index can be used to perform
time series segmentation: dividing a time series in in-
ternally consistent regimes (Gharghabi et al., 2017).
By examining the arcs defined by the matches found
by the Matrix Profile, they explain how a lower-than-
expected number of arcs at a specific timestamp gives
an indication of a change in the underlying system.
Their FLUSS algorithm and its online variant FLOSS
are found on average to outperform human segmen-
tation, based on experiments with 22 subjects and 12
data sets.
The Matrix Profile has been used for various tech-
niques and applied to data from various domains.
However, data where flat and noisy subsequences are
present has been mostly avoided in Matrix Profile re-
lated literature, most likely due to the issue introduced
in Section 1. In fact, we suspect this issue will nega-
tively affect many applications of the Matrix Profile,
severely limiting its applicability. To the best of our
knowledge, no technique has been published so far
to tackle the negative effects of flat and noisy subse-
quences. In the following section, we will elaborate
on this issue and provide a solution for it.
3 ELIMINATING NOISE
In this section, we present our contribution: a way to
remove the effects of noise from the Matrix Profile. In
Section 3.1, we first estimate the effect of noise when
calculating the z-normalized Euclidean distance be-
tween identical sequences. Next, we present how to
use this estimate in the calculation of the Matrix Pro-
file in Section 3.2. Finally, we discuss the complexity
of the resulting algorithm in Section 3.3.
3.1 Estimating the Influence of Noise
In this section we will estimate the amount of noise
affecting the Matrix Profile using statistical methods.
By taking this estimate into account while calculating
the Matrix Profile, we will be able to greatly reduce
the influence of noise.
Assume we have a base sequence S
S
S ∈ R
m
and de-
rive two sequences X
X
X and Y
Y
Y by adding Gaussian noise
N
N
N
1
and N
N
N
2
that was sampled from a normal distribu-
tion with variance σ
2
N
.
S
S
S = (s
s
s
1
, s
s
s
2
, . . . s
s
s
m
)
X
X
X = (s
s
s
1
+ n
n
n
1
1
, s
s
s
2
+ n
n
n
1
2
, . . . s
s
s
m
+ n
n
n
1
m
)
Y
Y
Y = (s
s
s
1
+ n
n
n
2
1
, s
s
s
2
+ n
n
n
2
2
, . . . s
s
s
m
+ n
n
n
2
m
)
Ideally, we would want the Euclidean distance be-
tween X
X
X and Y
Y
Y to be zero, since they are both (noisy)
measurements of the exact same sequence S
S
S. Remem-
ber that the Matrix Profile uses the Euclidean distance
of the z-normalized sequences.
D(X
X
X, Y
Y
Y ) = EucDist(
ˆ
X
X
X,
ˆ
Y
Y
Y )
= EucDist
X
X
X −µ
X
X
X
σ
X
X
X
,
Y
Y
Y − µ
Y
Y
Y
σ
Y
Y
Y
=
q
(
ˆ
x
x
x
1
−
ˆ
y
y
y
1
)
2
+ . . . + (
ˆ
x
x
x
m
−
ˆ
y
y
y
m
)
2
(1)
Here,
ˆ
X
X
X denotes the z-normalized sequence, µ de-
notes the mean and σ the standard deviation.
We now determine the expected influence of the
noise. From here on, we consider the sequences as
random variables, and highlight this by referencing
them as X and Y .
E
D(X, Y )
2
= E
( ˆx
1
− ˆy
1
)
2
+ . . . + ( ˆx
m
− ˆy
m
)
2
= m · E
( ˆx − ˆy)
2
= m · E
"
x − µ
X
σ
X
−
y − µ
Y
σ
Y
2
#
(2)
Since X and Y are the sum of the same two uncor-
related variables, they both have the same variance.
σ
2
X
= σ
2
Y
= σ
2
S
+ σ
2
N
(3)
Furthermore, we can decompose µ
X
and µ
Y
in the
original µ
S
and the influence of the noise. Here we
use n as the random variable sampled from the noise
distribution. Note that µ
S
can be seen as a constant.
Eliminating Noise in the Matrix Profile
85
µ
X
= µ
Y
= µ
S
+
n
1
+ . . . + n
m
m
= µ
S
+ µ
N
µ
N
∼ N
0,
σ
2
N
m
(4)
We can do the same for x and y, where s is an
unknown constant:
x = y = s + n
n ∼ N
0, σ
2
N
(5)
Using (3), (4) and (5) in (2), canceling out con-
stant terms and merging the distributions results in:
E
D(X, Y )
2
= m · E
n
x
− n
y
− µ
N
x
+ µ
N
y
q
σ
2
S
+ σ
2
N
2
= m · E
h
(ν)
2
i
ν ∼ N
0,
2 + 2m
m
·
σ
2
N
σ
2
S
+ σ
2
N
(6)
Finally, we apply the theorem E[X
2
] = var(X) +
E[X]
2
:
E
D(X, Y)
2
= (2m + 2) ·
σ
2
N
σ
2
S
+ σ
2
N
(7)
We now have an estimate of the distance between
two sequences that originate from the same sequence
but have been contaminated by Gaussian noise. Note
that in (7), σ
2
N
is the variance of the noise and σ
2
S
+σ
2
N
is the variance of the noisy sequence.
3.2 Algorithm for Noise Elimination
Implementing noise elimination is straightforward,
we subtract the squared estimate from the original
squared distance (e.g. as calculated by the MASS al-
gorithm). We do this before the element-wise mini-
mum is calculated and stored as value for the Matrix
Profile, because this correction might influence which
subsequence gets chosen as the best match. Pseu-
docode is listed below.
Input: d # distance between subsequences X, Y
Input: m # length of each subsequence
Input: std_X # standard deviation of X
Input: std_Y # standard deviation of Y
Input: std_n # standard deviation of noise
d_corrected = sqrt(dˆ2 - (2 + 2m) * std_nˆ2 /
max(std_X, std_Y)ˆ2)
return d_corrected
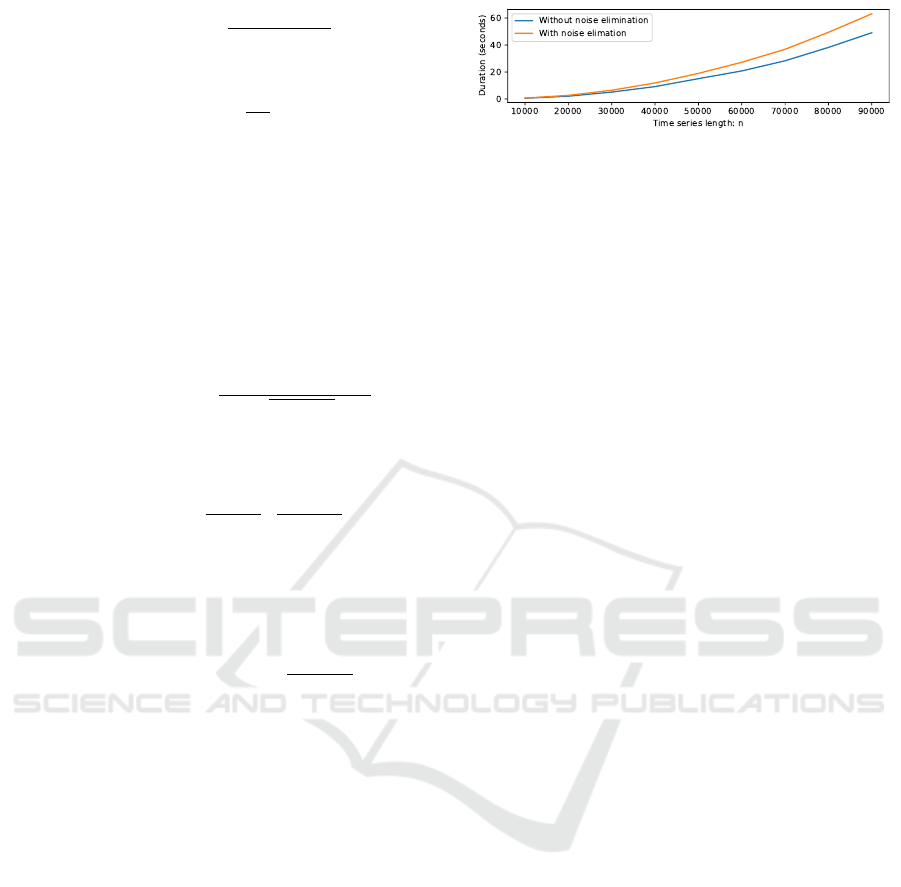
Figure 2: Time measurements show the overhead of the
noise elimination, though the complexity remains O(n
2
).
Note that the maximum of the standard deviation
of both sequences is used. Given two fundamentally
different subsequences, this choice effectively mini-
mizes our estimate. We note that we tried other means
of combining the standard deviations such as mean
and minimum, but these produced similar results.
3.3 Complexity Analysis
The noise elimination is a constant operation if we
know the standard deviations, so complexity remains
O(n
2
logn) when using STAMP and O(n
2
) when us-
ing STOMP. This assumes the standard deviations for
all subsequences are precomputed, as is the case in
the STOMP and STAMP algorithms. A plot of the in-
fluence of time series length versus runtime is shown
in Figure 2.
4 USE CASE: ANOMALY
DETECTION
One of the original applications for the Matrix Pro-
file was finding discords. Since discords are defined
as the subsequences in a series that differ most from
any other subsequence, they can be interpreted as
anomalous subsequences. Since each Matrix Profile
value represents the distance of each subsequence to
its nearest match, the top discord can be found triv-
ially given the Matrix Profile: it is the subsequence
corresponding to the highest value. When interested
in the top-k discords, one can take the top-k values
of the Matrix Profile where each value should be at
least m index positions away from all previous dis-
cord locations. Since overlapping subsequences are
very similar in shape, this prevents selecting the same
discord multiple times (Mueen et al., 2009).
In this section, we will use anomaly detection as
use case to demonstrate the influence of noise on the
Matrix Profile. We first focus on a small synthetic
dataset to provide details and insights, after which we
present results for real-world data from the Numenta
Anomaly Benchmark (Lavin and Ahmad, 2015).
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
86
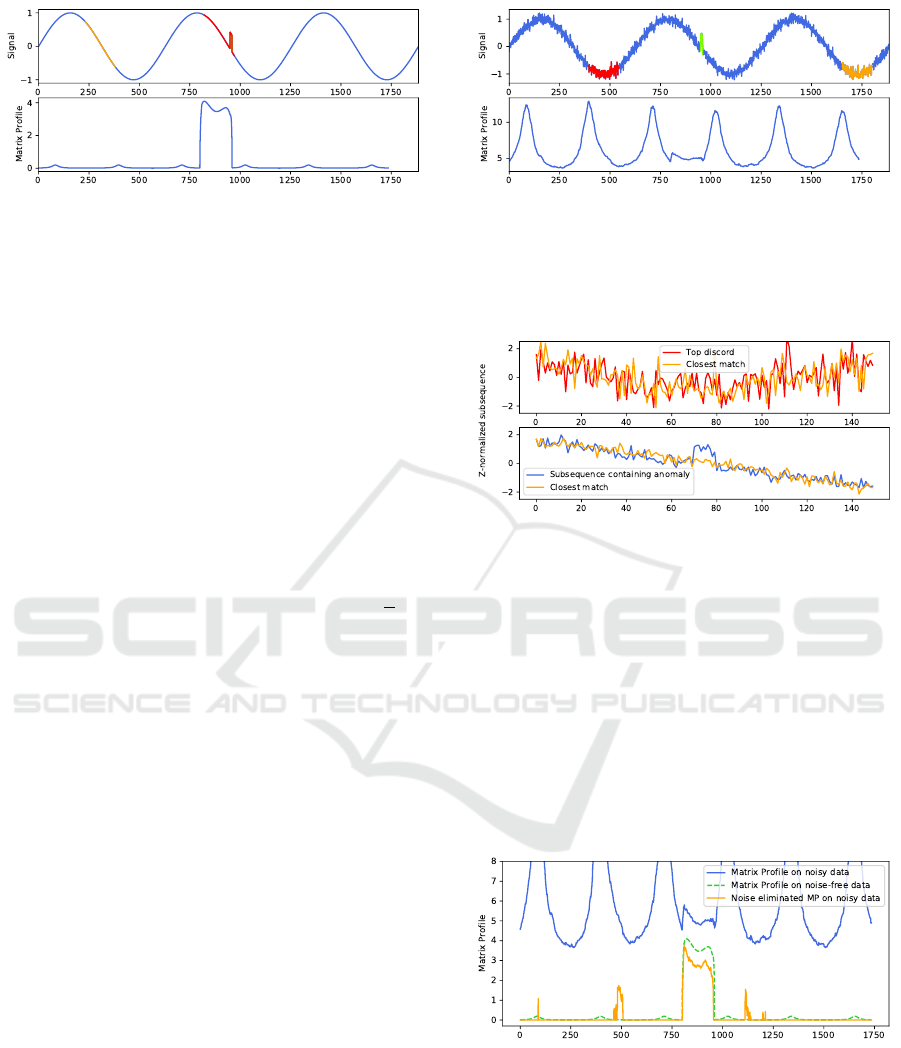
Figure 3: Top: Sinusoid signal, an anomaly of length 10
was added at index 950. The top discord (containing the
anomaly) and its closest match are highlighted in red and
orange. (The top discord subsequence starts at the highest
value of the Matrix Profile.) Bottom: Corresponding Matrix
Profile calculated with subsequence length m = 150. The
effect of the anomaly stands out as a series of high values.
4.1 Synthetic Data
We start from a synthetic dataset of 2000 samples of
a sinusoid in which we introduced an anomaly in one
of the downward slopes by increasing the value of 10
consecutive samples by 0.5. The dataset and corre-
sponding Matrix Profile can be seen in Figure 3. Here,
we see how the Matrix Profile clearly highlights the
subsequences containing the anomaly. For calculat-
ing the Matrix Profile, we used a subsequence dis-
tance m of 150 and a trivial match buffer of
m
2
, as
recommended in (Yeh et al., 2016).
Next, we added Gaussian noise sampled from
N (0, 0.01) and recalculate the Matrix Profile using
the same parameters. The resulting dataset and cor-
responding Matrix Profile can be seen in Figure 4.
Although we see still the impact of the anomaly in
the Matrix Profile as a deviation in the pattern, it
does not lead to a high value in the Matrix Profile.
In a larger, more realistic dataset, this could mean
that the anomaly would go unnoticed. Instead, the
top-6 discords coincide with the flat sections of the
sinusoid, where the noise has been upscaled due to
the z-normalization. This is visualized in Figure 5,
where we overlay the z-normalizations of the subse-
quences of the top discord and its best match with a
subsequence containing the data anomaly and its best
match. Since the Matrix Profile consists of the Eu-
clidean distance between z-normalized subsequences,
more erratic z-normalized subsequences will typically
lead to higher distances.
After recalculating the Matrix Profile using our
noise elimination technique from Section 3, the Ma-
trix Profile again closely matches the Matrix Profile
on the noise-free data, as can be seen in Figure 6.
Most subsequences now have a distance of zero, in-
dicating exact matches in the remainder of the se-
ries. The anomaly is again clearly visible and would
be marked as the top discord. Lastly, we see some
Figure 4: Top: Same signal as Figure 3, but with added
Gaussian noise. The anomaly is highlighted in green. The
top discord and its closest match are highlighted in red and
orange, they do not contain the data anomaly. Bottom: The
corresponding Matrix Profile, the effect of the anomaly is
still visible but no longer stands out as a high value.
Figure 5: A close-up of the z-normalized subsequences
explains why the top discord has a higher Matrix Profile
value than any subsequence containing the data anomaly.
The effect of the noise becomes larger due to the z-
normalization when the original subsequence is “flatter”.
Top: z-normalized subsequence of the top discord and
its closest match (as marked in Figure 4). Bottom: z-
normalized subsequence that contains the data anomaly and
its closest match.
non-zero values not related to the anomaly, these are
caused by locally higher-than-expected noise values
in that part of the signal. It can be noted that these
values depend on the sampling of the noise and can in
fact be seen as subtle anomalies.
Figure 6: Plot of the Matrix Profile for the noisy data (blue),
for noise-free data (dashed green) and for the noisy data, but
with the noise elimination applied (orange). We see that the
noise corrected Matrix Profile closely matches the Matrix
Profile of the noise-free data.
Let us briefly further investigate how the proper-
ties of the noise affect the Matrix Profile. Figure 7
displays our starting sinusoidal signal and anomaly,
to which Gaussian noise sampled from different dis-
Eliminating Noise in the Matrix Profile
87
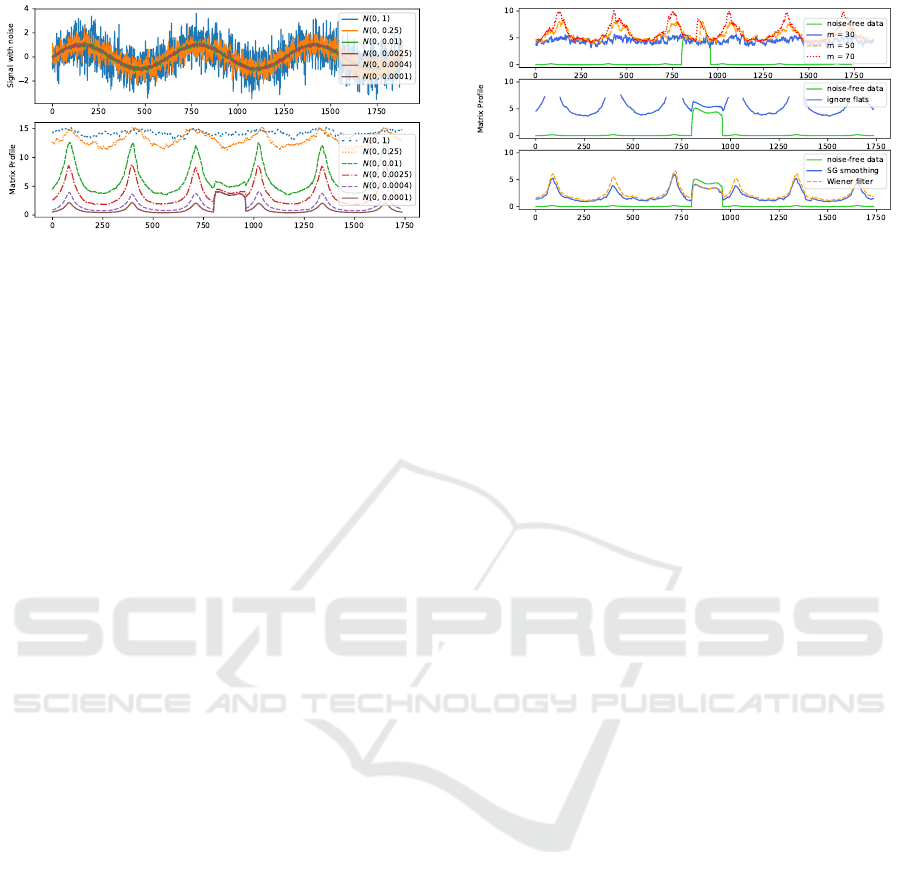
Figure 7: Top: Sinusoid signal with anomaly to which
Gaussian noise sampled from different distributions has
been added. Bottom: Corresponding Matrix Profile cal-
culated with subsequence length m = 150 for each of the
noisy signals. We see how the Matrix Profile becomes less
insightful as the amount of noise increases.
tributions was added. As expected, we see that as the
variation of the noise increases, the anomaly becomes
less apparent in the Matrix Profile. The anomaly is no
longer visually obvious in the Matrix Profile for noise
with variance of 0.25 or more. Somewhat surprising
is how quickly this effect becomes apparent: when the
noise has a variance of around 0.0004 (at this point the
signal-to-noise ratio is 1250 or 31 dB), the anomaly is
already occasionally overtaken as the top discord by
the flat subsequences (depending on the sampling of
the noise). The noise cancellation technique becomes
unreliable once the variation is around 0.0625, and is
no longer useful for variations of 0.25 or higher. This
can be explained by the subtlety of our anomaly (an
increase by 0.5 for 10 time units), as high variance
noise can create similar artifacts. Note that these in-
sights are hard to generalize as a rule of thumb, as they
really depend on the properties of the time series, the
anomalies and the considered subsequence length.
Lastly, we discuss why some simple, seemingly
useful methods to tackle the effects of noise are not
generally usable.
• Changing the subsequence length m: as m be-
comes smaller, the effect of the anomaly on the
Matrix Profile will indeed be bigger. However,
the effect of the noise also becomes bigger, result-
ing in a more eratic Matrix Profile, still making it
hard to find the actual anomaly. Without knowing
in advance what one is looking for, it is hard to
select a good value of m. This is demonstrated in
Figure 8 (top).
• Ignoring flat sections: ignoring subsequences
whose variance is below a certain value would
result in removing the peaks in the Matrix Pro-
file. A first problem is finding the correct cut-
off value, which is not trivial. Secondly, this ap-
proach would not be applicable in datasets where
flat signals are the anomalies, or where matches
Figure 8: Approaches for negating the effects of noise that
do not work. Top: Matrix Profile using different values for
the subsequence length m. Middle: Matrix Profile while
ignoring subsequences with standard deviation below 0.2.
Bottom: Matrix Profile after applying a Savitzky-Golay
smoothing and a Wiener frequency filter to the noisy data.
on flat signals are useful, as is demonstrated in
the time series segmentation of Section 5. This
approach is demonstrated in Figure 8 (middle).
• Smoothing or filtering: by preprocessing the noisy
signal, one could hope to remove the noise alto-
gether. Unfortunately, unless the specifics of the
noise are well known and the noise can be com-
pletely separated from the signal, there will al-
ways remain an amount of noise, resulting in the
same effects. This is demonstrated in Figure 8
(bottom).
4.2 Numenta Anomaly Benchmark
The Numenta Anomaly Benchmark (NAB) is a col-
lection of datasets, score metrics and supporting tools.
Most datasets are from real-world applications in dif-
ferent domains and have been annotated by hand
by multiple people and combined using a consen-
sus method. The benchmark is aimed at real-time
anomaly detectors, this is reflected in the scoring
method, where anomaly time windows are defined and
detection in the start of the window is better rewarded.
We limited ourselves to the “realAWSCloud-
watch” collection, because this collection was the
only one that contained many flat, noisy time series.
The collection consists of 17 datasets of real-world
data, measuring computer workload such as cpu uti-
lization, bytes written and request count. Each dataset
spans a time period of 4 to 16 days sampled at 5
minute intervals and contains 0 to 3 anomalies. One
dataset with 2 anomalies is visualized in Figure 9.
We did not use the score metric of the NAB, as
it requires labelling anomalies in a streaming fash-
ion, which would require optimizing a classification
threshold for the Matrix Profile. Instead, we relied
on the inherent ordering of normal to anomalous sub-
sequences that is captured in the Matrix Profile: We
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
88
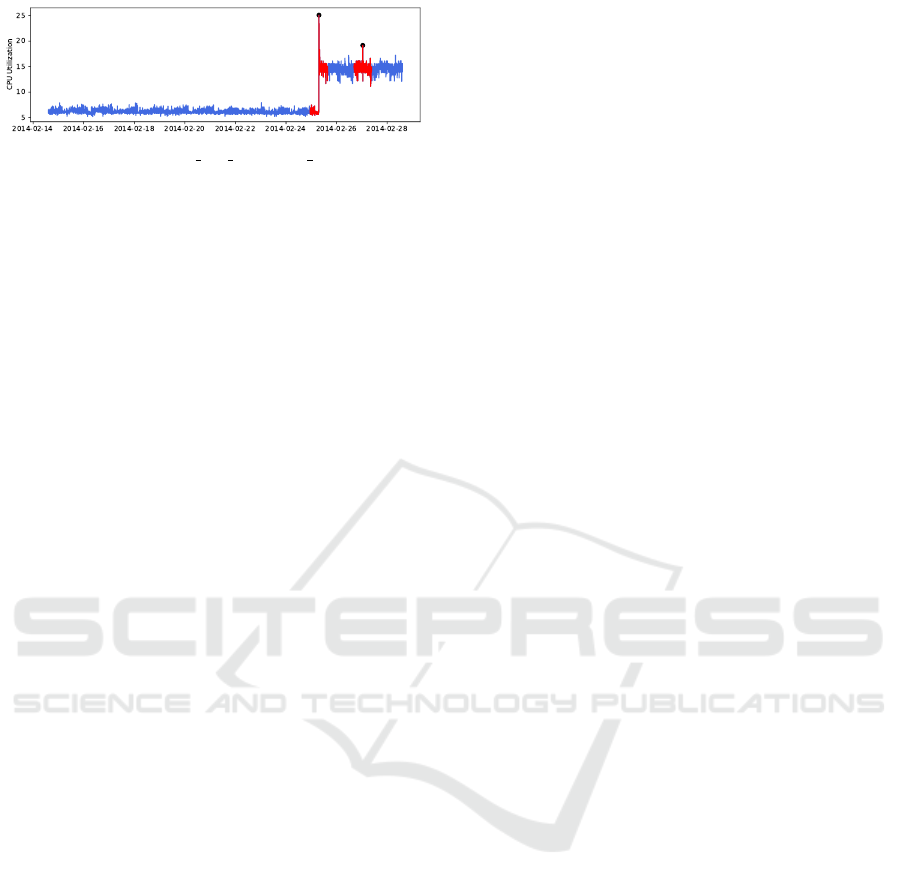
Figure 9: Plot of the “rds cpu utilization cc0c53” dataset
from the Numenta Anomaly Benchmark, it contains 2
anomalies (dots) with associated anomaly windows (red).
calculated the Matrix Profile over the time series and
counted how many guesses were needed to find all
anomalies, with a limit of 10 wrong guesses. Note
that when calculating the Matrix Profile, we only used
data from previous time steps to define normal behav-
ior, this is in line with the real-time anomaly detection
idea of the NAB and is needed to ensure we match
their definition of anomalous behavior.
We compare the performance of the Matrix Pro-
file with and without noise elimination. For each
dataset, we set the subsequence length equal to half
the anomaly time window (which is defined by the
NAB as 10% of the data length, divided by the num-
ber of anomalies present). Furthermore, like in the
NAB, we use the first 15% of the data as reference
data and do not report any anomalies in it. For the
noise elimination, we estimated the standard devia-
tion of the noise as the 5th percentile of the standard
deviations of all subsequences found in the reference
data. This value was similar to a manually estimated
value on a few test datasets and was selected as an
easy, automatically derivable value. Note that we did
not optimize the noise estimation value in any way.
Our results are listed in Table 1, where we see that
noise elimination has better results for 6 and worse re-
sults for 2 out of the 17 datasets. One of these worse
results can be explained because the reference data
contains a very noisy signal that later becomes more
stable, this causes an overestimation of the influence
of noise and results in exact matches being found for
the entire time series. More fine grained control of
the noise parameter could likely correct this case. In
total, by using noise elimination, we found 28 out 30
anomalies, and made 56 wrong guesses in the pro-
cess. In comparison, the original approach found 24
anomalies and had 80 wrong guesses. These results
show that by using the noise elimination, anomalies
become generally more noticeable and false positives
are reduced in the Matrix Profile, resulting in less time
lost by the experts who diagnose these reports.
5 USE CASE: SEGMENTING
TIME SERIES
Detecting changes in an underlying system based on
sensor measurements has a wide range of applica-
tions, such as identifying different speakers in an au-
dio recording, detecting system intrusion or data anal-
ysis in general. In this section we will perform time
series segmentation using the Corrected Arc Curve
(CAC), a technique based on the Matrix Profile. The
CAC is calculated by the FLUSS algorithm or its on-
line variant, the FLOSS algorithm, all are introduced
in (Gharghabi et al., 2017).
The CAC is coined as a domain agnostic tech-
nique to perform semantic time series segmentation
at super-human performance on realistic datasets and
even on streaming data. Both FLUSS and FLOSS
work by analyzing the Matrix Profile Index, which
contains the index of the closest match for each sub-
sequence in a time series. To calculate the CAC,
the number of arcs (each spanning between a subse-
quence and their respective best match) are counted
and divided by the expected amount of arcs if all
matches were determined by uniform sampling over
the entire time series. Assuming that homogeneous
segments will display similar behavior and heteroge-
neous segments will not, a low ratio is seen as evi-
dence of a change in the underlying system. The CAC
is a vector of the same length as the Matrix Profile and
is typically restricted to values between 0 and 1, the
lower the value, the more evidence of a change in the
underlying system.
5.1 Evaluation on PAMAP2 Dataset
The PAMAP2 dataset (Reiss and Stricker, 2012) con-
tains sensor measurements of 9 subjects performing
a subset of 18 activities including: sitting, standing,
walking, ironing and so on. The measurements are
time series containing the output of a heart rate mon-
itor and 3 inertial measurement units (IMU) placed
on the chest, dominant wrist and dominant ankle of
each subject. Each IMU measured 3-D acceleration
data, 3-D gyroscope data and 3-D magnetometer data
at around 100 Hz. The time series are annotated with
the activity being performed by the subject or marked
as a transition region in between activities. The dura-
tion of each activity varies greatly, but most activities
last between 3 to 5 minutes.
The PAMAP2 dataset was used in (Yeh et al.,
2017a), where the authors used the Matrix Profile to
classify the activities in passive and active activities.
Specifically, they note that the motif pairs in the pas-
sive actions are less similar and therefore less useful.
Eliminating Noise in the Matrix Profile
89
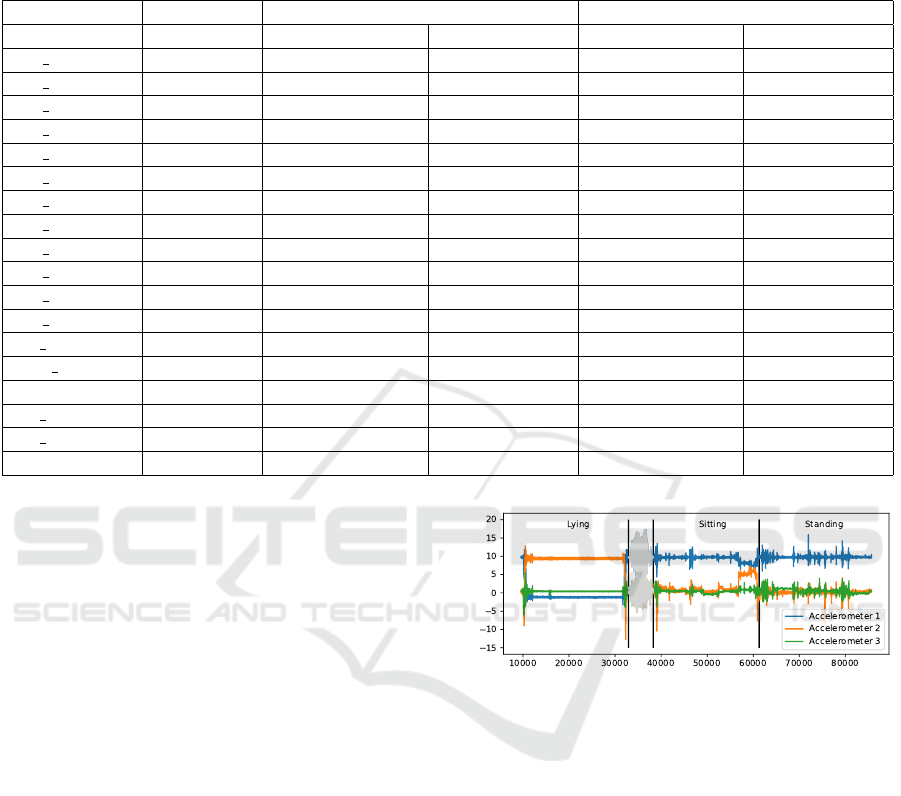
Table 1: Results of anomaly detection using the Matrix Profile with and without noise elimination on the “realAWSCloud-
watch” collection of the Numenta Anomaly Benchmark. Each model kept guessing until all anomalies were found or until
10 wrong guesses occurred. Bold entries indicate a better performance of one approach over the other. We see that by using
noise elimination we can more easily find the annotated anomalies in most cases.
Without Noise Elimination With Noise Elimination
Dataset #Anomalies Found Anomalies Wrong Guesses Found Anomalies Wrong Guesses
ec2 cpu...e8d 2 0 10 2 0
ec2 cpu...a38 2 2 6 2 0
ec2 cpu...533 2 2 1 2 10
ec2 cpu...1ca 1 0 10 1 9
ec2 cpu...cc2 1 1 0 1 0
ec2 cpu...0cd 1 1 1 1 0
ec2 cpu...85a 0 0 0 0 0
ec2 cpu...f93 3 1 10 1 10
ec2 disk...3de 1 1 3 1 3
ec2 disk...644 3 3 10 3 10
ec2 netw...a54 1 1 0 1 0
ec2 netw...ac7 2 2 3 2 3
elb reques... 2 2 10 2 0
grok asg... 3 3 3 3 3
ii...NetworkIn 2 2 0 2 5
rds cpu...c53 2 1 10 2 0
rds cpu...b3b 2 2 3 2 3
Sum 30 24 80 28 56
We argue that motifs in passive actions, consisting of
mainly flat signals, can also help us find structure in
the data if we can remove the influence of the noise.
We applied time series segmentation on the passive
activities present in the PAMAP2 dataset, focusing
on following activities: lying, sitting and standing.
We picked these activities since their measurements
display very few patterns in the data and they are con-
secutive activities for all subjects, meaning we did not
have to introduce time-jumps in the data.
We considered subjects 1 to 8 of the dataset (sub-
ject 9 has no recordings of the relevant activities) and
tested 2 cases: the transition from “lying” to “sit-
ting” and the transition from “sitting” to “standing”.
For each subject, we used the 3 accelerometer signals
from the IMU placed on the chest of the subject, any
missing data points were filled in using linear inter-
polation. We calculated the CAC with and without
noise elimination using a subsequence length of 1000
(10 seconds). The standard deviation of the noise was
estimated (without optimizing) by taking the 5th per-
centile of the standard deviations of all subsequences,
similar to our approach in Section 4.
An example of the signals can be seen in Figure
10. We emphasize it is not our goal to build the opti-
mal segmentation tool for this specific task, we simply
want to evaluate the effect of the noise elimination on
the CAC for sensor signals that contain flat but noisy
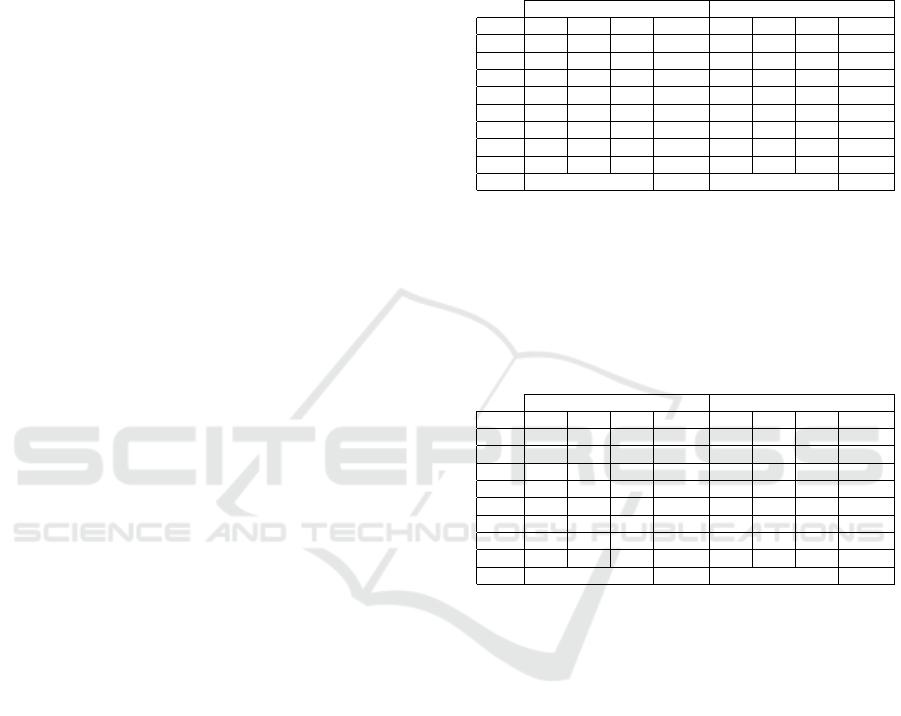
Figure 10: Three chest accelerometers of subject 6 from
the PAMAP2 dataset. This extract contains 3 activities and
one transition period. No clear patterns are discernible and
many flat and noisy subsequences are present.
subsequences.
There is one side effect of the noise cancellation
that needs attention before calculating the CAC. After
applying noise cancellation, many flat subsequences
will have exact matches to other flat subsequences.
However, since the Matrix Profile Index can only
store a single match, the calculation order determines
which match actually gets stored. This causes an un-
wanted pattern in the Matrix Profile indices, which
contradicts the assumption of the CAC that the struc-
ture of matches is related to similar segments. Note
that this effect is already present in the normal Matrix
Profile, but typically has little to no influence because
multiple exact matches are extremely rare.
To solve this, we introduced reservoir sampling
(Vitter, 1985) to the construction of the Matrix Profile
Index. Reservoir sampling allows uniform sampling
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
90
without replacement from a stream without knowing
the size of the stream in advance. We implemented
reservoir sampling so that the Matrix Profile Index no
longer stores the first encountered best match, but a
uniformly sampled instance of all matches with the
lowest distance. This required us to store an addi-
tional vector of the same length as the Matrix Pro-
file, keeping track of the number of exact matches that
was encountered so far for each subsequence. Pseudo
code to update the Matrix Profile and its indices is
listed below.
# Matrix Profile and Index being updated
Input: mp, mpi
# Distances (e.g. from STOMP iteration)
Input: dists
# Indices corresponding to dists
Input: indices
# Counts exact matches per subsequence
Input: match_counts
better_indices = dists < mp
eql_indices = dists == mp AND finite(dists)
mp(better_indices) = dists(better_indices)
mpi(better_indices) = indices(better_indices)
match_counts(better_indices) = 1
match_counts(eql_indices) += 1
for i in eql_indices:
if rand() < 1 / match_counts(i):
mpi(i) = indices(i)
return (mp, mpi)
For both test cases (lying-sitting and sitting-
standing), we calculated the CAC using the reser-
voir sampled Matrix Profile indices with and without
noise elimination for each individual sensor. From the
CAC, we derived a single activity-transition point by
taking the location where the CAC is minimal. We
considered 4 segmentations: one for each CAC of the
3 sensor channels and one obtained by combining (av-
eraging) these individual CACs.
For scoring, we wish to reward splits on or near
the transition periods as annotated in the data sets.
Note that some transitions are instantaneous, while
others consist of a transient period, as can be seen in
Figure 10. We added a buffer margin of subsequence
size m to the transitions, so each transition period is at
least 2m wide. As score, we took the distance between
the estimated transition location and the buffered tran-
sition period, normalized by the length of the series
(containing both activities and the transition period).
Pseudo code for this scoring function is listed below,
a score will range from 0 to 100, where lower is better.
Table 2: Score for the segmentation of the transition from
“lying” to “sitting” using the 3 chest accelerometers from
the PAMAP2 dataset for subjects 1 through 8, with and
without noise elimination applied. Segmentation is per-
formed using the CAC from a single sensor (C1, C2 and
C3) and using the combined CAC. We see similar or better
performance when applying noise elimination for all sub-
jects except subject 1.
Without NE With NE
Subj. C1 C2 C3 Comb. C1 C2 C3 Comb.
1 5.9 31.3 31.9 31.7 41.3 31.8 41.8 36.7
2 32.9 1.4 1.4 1.4 28.8 1.4 1.7 1.4
3 35.9 2.8 31.1 33.8 2.4 2.3 2.3 2.3
4 0.0 2.8 5.9 0.0 0.0 1.5 6.6 0.8
5 1.1 7.6 5.1 3.9 1.6 1.7 4.9 1.6
6 2.5 1.9 2.3 2.3 2.4 1.9 2.0 2.4
7 0.1 1.8 11.1 2.0 2.1 1.8 1.9 1.9
8 0.0 1.4 5.5 1.7 0.0 1.4 1.4 1.4
Avg. 9.32 9.61 7.71 6.07
Table 3: Score for the segmentation of the transition from
“sitting” to “standing” using the 3 chest accelerometers
from the PAMAP2 dataset for subjects 1 through 8, with
and without noise elimination applied. Segmentation is per-
formed using the CAC from a single sensor (C1, C2 and
C3) and using the combined CAC. Overall, we see similar
or better performance when applying noise elimination, ex-
cept for the segmentation using the first channel for subject
1, 3 and 8.
Without NE With NE
Subj. C1 C2 C3 Comb. C1 C2 C3 Comb.
1 32.5 0.0 3.6 2.2 38.7 0.0 3.7 2.2
2 36.5 37.2 36.4 37.0 7.1 30.0 32.7 29.2
3 10.0 30.2 43.1 30.2 43.2 14.0 43.7 30.2
4 7.8 1.9 1.1 1.2 0.7 2.0 1.3 1.3
5 13.1 0.0 28.5 10.6 13.3 1.0 1.2 1.0
6 36.1 36.6 26.9 36.6 23.3 3.4 26.5 3.2
7 43.1 38.0 16.5 16.5 43.4 1.6 0.0 1.6
8 2.3 1.0 24.8 1.0 21.1 0.0 16.5 1.0
Avg. 21.12 16.9 15.35 10.3
Input: est_split # Algorithmic estimate
Input: split_start, split_end # Ground truth
Input: n # Length of the time series
Input: b # Buffer for transition
if est_split < split_start - b:
return ((split_start - b) - est_split)/n
elif est_split > split_end + b:
return (est_split - (split_end + b))/n
else:
return 0.
The results for segmentation on the transition
from “lying” to “sitting” are listed in Table 2. We see
that segmentation using the individual sensors as well
as the combined approach provides similar or better
results when using noise elimination for all subjects
except for subject 1. The average score for the indi-
vidual sensors improves from 9.32 to 7.71, a modest
improvement corresponding to a gain of about 8 sec-
onds. The segmentation based on all 3 sensor series
improves from 9.61 to 6.07, a gain of about 18.5 sec-
onds. The results for subject 1 can be attributed to
Eliminating Noise in the Matrix Profile
91

changes of the subjects body position near the start
of the “lying” activity. This results in a lower-than-
expected amount of matches near the start, causing
the transition to be guessed too early. Note that both
techniques have trouble with subject 1.
Table 3 lists the results for the transition from
“sitting” to “standing”. Again, we see similar or
improved results when applying noise elimination,
except for 3 subjects using the first sensor series.
Though results are worse compared to Table 2, the
gain by using noise elimination is more significant.
When using a single sensor, results on average im-
prove from 21.12 to 15.35, corresponding to a gain of
about 27 seconds. If segmentation uses all 3 sensors,
results change from 16.9 to 10.3 on average, a gain of
around 31 seconds.
6 CONCLUSION
In this paper we discussed techniques and applica-
tions related to the Matrix Profile. Our contribution
consists of a method to remove the effects of Gaussian
noise on the time series when calculating the Matrix
Profile, without affecting the complexity of the un-
derlying algorithm. This method is based on a sta-
tistical analysis of the effects of z-normalized noisy
sequences on the Euclidean distance. The only re-
quirement for this technique is to know the variance
of the noise, which is an intuitive measure and can
be easily estimated by manually extracting a flat but
noisy segment from the time series.
As the Matrix Profile is widely usable for a va-
riety of problems and across various domains, so
is our technique. In this paper, we showed gains
for anomaly detection and time series segmentation.
Both cases were evaluated on public datasets contain-
ing real-word data and showed an improvement of
the results. On the Numenta Anomaly Benchmark,
we were able to retrieve more anomalies with less
guesses, saving an operator valuable time. On the
PAMAP2 dataset, we were able to more accurately
predict transitions between passive activities. We fo-
cused on datasets containing flat and noisy segments,
a subject that was not yet tackled in other Matrix Pro-
file related literature.
Further work can be done on using a dynamic
value for the noise estimation, for series where noise
does not originate from measurement, but rather from
the underlying process.
ACKNOWLEDGEMENTS
This work has been carried out in the framework
of the Z-BRE4K project, which received funding
from the European Union’s Horizon 2020 research
and innovation programme under grant agreement no.
768869.
REFERENCES
Dau, H. A. and Keogh, E. (2017). Matrix Profile V: A
Generic Technique to Incorporate Domain Knowledge
into Motif Discovery. In Proceedings of the 23rd
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining - KDD ’17, pages
125–134, New York, New York, USA. ACM Press.
Gharghabi, S., Ding, Y., Yeh, C.-C. M., Kamgar, K.,
Ulanova, L., and Keogh, E. (2017). Matrix Pro-
file VIII: Domain Agnostic Online Semantic Seg-
mentation at Superhuman Performance Levels. In
2017 IEEE International Conference on Data Mining
(ICDM), pages 117–126. IEEE.
Keogh, E. and Kasetty, S. (2002). On the need for time
series data mining benchmarks. Proceedings of the
eighth ACM SIGKDD international conference on
Knowledge discovery and data mining - KDD ’02,
page 102.
Lavin, A. and Ahmad, S. (2015). Evaluating Real-
Time Anomaly Detection Algorithms – The Numenta
Anomaly Benchmark. In 2015 IEEE 14th Interna-
tional Conference on Machine Learning and Applica-
tions (ICMLA), pages 38–44. IEEE.
Mueen, A., Keogh, E., Zhu, Q., Cash, S., and Westover, B.
(2009). Exact Discovery of Time Series Motifs. In
Proceedings of the 2009 SIAM International Confer-
ence on Data Mining.
Mueen, A., Viswanathan, K., Gupta, C., and Keogh,
E. (2015). The fastest similarity search al-
gorithm for time series subsequences under eu-
clidean distance. url: www. cs. unm. edu/˜
mueen/FastestSimilaritySearch. html (accessed 24
May, 2016).
Papadimitriou, S. and Faloutsos, C. (2005). Streaming Pat-
tern Discovery in Multiple Time-Series. International
Conference on Very Large Data Bases (VLDB), pages
697–708.
Reiss, A. and Stricker, D. (2012). Introducing a new bench-
marked dataset for activity monitoring. In Proceed-
ings - International Symposium on Wearable Comput-
ers, ISWC, pages 108–109.
Vandewiele, G., Colpaert, P., Janssens, O., Van Herwe-
gen, J., Verborgh, R., Mannens, E., Ongenae, F., and
De Turck, F. (2017). Predicting train occupancies
based on query logs and external data sources. In Pro-
ceedings of the 7th International Workshop on Loca-
tion and the Web.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
92

Vitter, J. S. (1985). Random sampling with a reser-
voir. ACM Transactions on Mathematical Software
(TOMS), 11(1):37–57.
Wang, X., Lin, J., Patel, N., and Braun, M. (2016). A
Self-Learning and Online Algorithm for Time Series
Anomaly Detection, with Application in CPU Manu-
facturing. In Proceedings of the 25th ACM Interna-
tional on Conference on Information and Knowledge
Management - CIKM ’16, pages 1823–1832, New
York, New York, USA. ACM Press.
Yeh, C.-C. M., Kavantzas, N., and Keogh, E. (2017a). Ma-
trix Profile VI: Meaningful Multidimensional Motif
Discovery. In 2017 IEEE International Conference
on Data Mining (ICDM), pages 565–574. IEEE.
Yeh, C. C. M., Van Herle, H., and Keogh, E. (2017b). Ma-
trix profile III: The matrix profile allows visualization
of salient subsequences in massive time series. Pro-
ceedings - IEEE International Conference on Data
Mining, ICDM, pages 579–588.
Yeh, C.-C. M., Zhu, Y., Ulanova, L., Begum, N., Ding, Y.,
Dau, H. A., Silva, D. F., Mueen, A., and Keogh, E.
(2016). Matrix Profile I: All Pairs Similarity Joins
for Time Series: A Unifying View That Includes Mo-
tifs, Discords and Shapelets. In 2016 IEEE 16th Inter-
national Conference on Data Mining (ICDM), pages
1317–1322. IEEE.
Zhu, Y., Imamura, M., Nikovski, D., and Keogh, E. (2017).
Matrix Profile VII: Time Series Chains: A New Prim-
itive for Time Series Data Mining (Best Student Paper
Award). In 2017 IEEE International Conference on
Data Mining (ICDM), pages 695–704. IEEE.
Zhu, Y., Zimmerman, Z., Senobari, N. S., Yeh, C.-c. M.,
Funning, G., Brisk, P., and Keogh, E. (2016). Matrix
Profile II : Exploiting a Novel Algorithm and GPUs
to Break the One Hundred Million Barrier for Time
Series Motifs and Joins. 2016 {IEEE} 16th Interna-
tional Conference on Data Mining ({ICDM}), pages
739–748.
Eliminating Noise in the Matrix Profile
93
