
Detecting a Fetus in Ultrasound Images using Grad CAM and
Locating the Fetus in the Uterus
Genta Ishikawa
1
, Rong Xu
2
, Jun Ohya
1
and Hiroyasu Iwata
1
1
Department of Modern Mechanical Engineering, Waseda University,3-4-1, Ookubo, Shinjuku-Ku, Tokyo, Japan
2
Global Information and Telecommunication Institute, Waseda University,3-4-1, Ookubo, Shinjuku-Ku, Tokyo, Japan
Keywords: Fetal, Ultrasound Image, Deep Leaning, Grad_CAM, Fetal Position.
Abstract: In this paper, we propose an automatic method for estimating fetal position based on classification and
detection of different fetal parts in ultrasound images. Fine tuning is performed in the ultrasound images to
be used for fetal examination using CNN, and classification of four classes "head", "body", "leg" and "other"
is realized. Based on the obtained learning result, binarization that thresholds the gradient of the feature
obtained by Grad Cam is performed in the image so that a bounding box of the region of interest with large
gradient is extracted. The center of the bounding box is obtained from each frame so that the trajectory of the
centroids is obtained; the position of the fetus is obtained as the trajectory. Experiments using 2000 images
were conducted using a fetal phantom. Each recall ratiso of the four class is 99.6% for head, 99.4% for body,
99.8% for legs, 72.6% for others, respectively. The trajectories obtained from the fetus present in “left”,
“center”, “right” in the images show the above-mentioned geometrical relationship. These results indicate that
the estimated fetal position coincides with the actual position very well, which can be used as the first step
for automatic fetal examination by robotic systems.
1 INTRODUCTION
Recently, ultrasonic images are frequently used for
fetal growth evaluation (Hadlock. R, 1983),
pregnancy duration estimation (Hadlock. R, 1981),
fetal weight estimation (Gull. I, 2002) etc. because of
safety, low cost, and impermeability. Among them,
weight estimation is one of the biometric
measurements that makes it easiest to evaluate the
growth of the fetus by measuring three parameters,
namely pediatric biparietal diameter (BPD),
abdominal circumference length (AC), and femur
length (FL) (Shinzi. T, 2014).
However, due to the discontinuity and irregularity of
the fetal head skull, low resolution and signal-to-
noise ratio in ultrasound images, the procedure of
fetal head detection depends on the physician's
experience and can be time consuming. On the other
hand, due to the shortage of doctor and gynecology, a
fully automated ultrasonic inspection robot system is
required.
Thus, the goal of this research is to develop a fully
automatic ultrasonic examination system for fetal
examination in pregnancy by robotics and image
processing. As a preliminary step, this paper aims at
identifying the ultrasound images required for the
examination and estimating fetal position by a series
of ultrasound images, where the fetal position means
the geometrical relationship between the fetus and the
uterine cavity.
Concerning fetal ultrasound examination, to classify
and localize cross sections of ultrasound images,
Sono_Net(Christian F, 2017) was developed by
Christian et al., and a weakly supervised localization
for fetal ultrasound images was developed by Nicolas
et al. (Nicolas T. 2018). Each sample image is
annotated by giving a class label, and their methods
learn the labelled data, so that the region of interest of
the class in each test image can be localized. In these
studies, medical doctors did the labelling task.
Medical doctors focus on BPD, AC, and FL, but not
edges of each part. In contrast, this paper focuses on
extracting edges of each fetal part. Also, there is no
research which estimates the fetal position in the
uterus. Yuanwei conducted a research to find BPD
from 3D ultrasound voxels (Yuanwei. L, 2018).
However, it is still difficlut to estimate the fetal
position in the uteras.
Our proposed approach is as follows. By learning the
features of the classes (head, body, legs, etc.), which
Ishikawa, G., Xu, R., Ohya, J. and Iwata, H.
Detecting a Fetus in Ultrasound Images using Grad CAM and Locating the Fetus in the Uterus.
DOI: 10.5220/0007385001810189
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 181-189
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
181
non-experts (not medical doctors) can easily locate
and identify, each class’ features, which for non-
experts it is hard to distinguish, are obtained, so that
the objects that correspond to each class can be
located.
The rest of this paper is organized as follows. Section
2 summarizes the data used in experiments. Section 3
explains the proposed algorithm. Section 4 presents
experimental results and discussion. Section 5
concludes the paper.
2 DATA
2.1 Image Data
The image data used in this paper have been accepted
by the ethical review. Image data was created by
extracting all frames from a 30 fps video. Regarding
the video, we use the videos in which a skilled
medical doctor performed ordinary examinations
manually. The videos contain 12 fetal data from the
16
th
to 34
th
pregnancy week. This pregnancy week
range is important for the ultrasonic examination like
the weight measurement, birth and fetal orientation.
Approximately 20,000 frames are extracted from
each video. The size of the image (frame) is 720 * 480
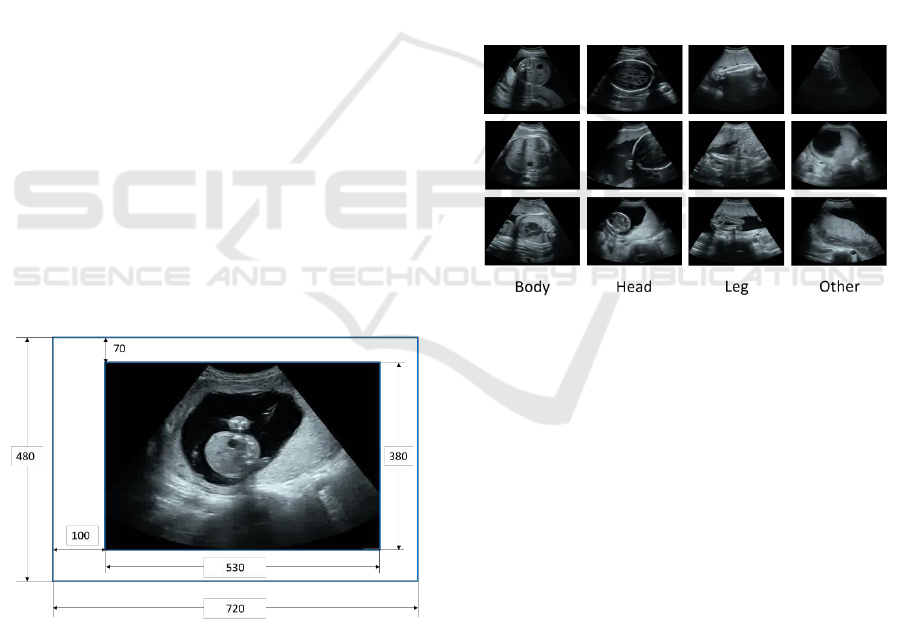
(pixels). Note that in this research, in order to hide the
patients’ personal information and examination
information, as shown in Fig. 1, only the ultrasound
image part is extructed as ROI (region of interest) and
used for the experiments.
Figure 1: Original ultrasound image and ROI.
2.2 Image Labels
We assigned four classes (head, body, legs, and
others) for all the ultrasound images. Examples of the
four classes are shown in Fig. 2. For non-experts, the
three classes: head, body and leg are easy to identify
and locate; that’s why we assign the four classes (the
three classes + others). Though the class of each
image is assigned by non-experts, the guidance of the
doctors and related reference (Shinzi. T, 2014) helps us
make acceptable annotations.
Head
The frame in which the skull is certainly visible as
well as its previous and next frames.
The head top is observed as a small circle.
Body
The frame in which the gastrocytes are visible in
circles with almost uniform gray and the spine exists
as well as its previous and next frames.
leg
There is a thick, straight white line. The pelvis and
two legs are visible.
Other
Other cannot be classified by the above-mentioned
three classes.
Figure 2: Criteria for classification.
Also, images in which multiple objects appear are
eliminated from the training images.
Totally, 21,110 frames with the head label, 23,300
with the body label, and 14,887 with the leg label.
Based on these data, we used 10,000 frames for
training and others for testing. From the training
frames, 2,000 frames are used for validation.
2.3 Data Augmentation
We expand the number of frames using data
augmentation for the images labelled in Section 2.2.
Types of augmentation are: moving to the left / right
direction, enlarging / reducing (Oquab..M, 2014),
(Razavian. A, 2014). Rotation and color tone
correction etc. are not carried out. Horizontal
movement, enlargement / reduction are performed
randomly, so that the number of deta is increased
about five times as many as the original frame number.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
182
3 ALGORITHM
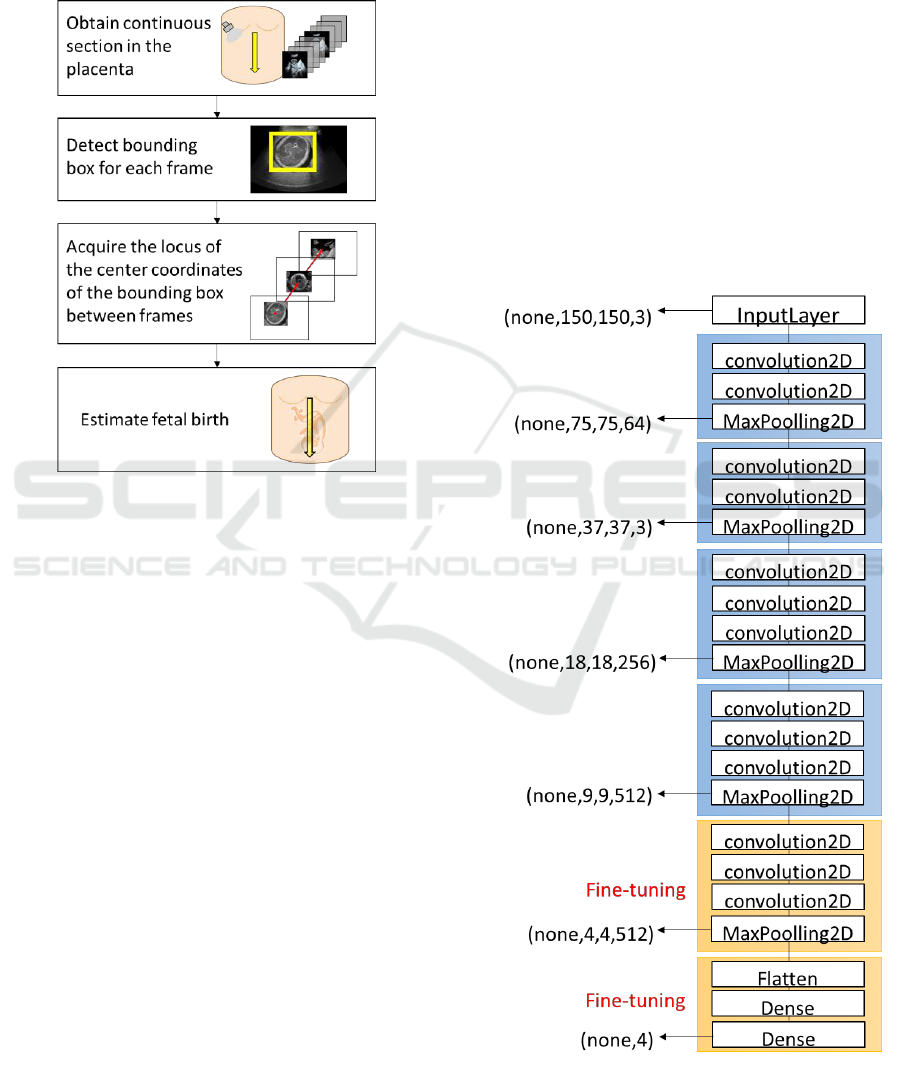
3.1 Overview
This section overviews the proposed method.
Figure 3: Overview of our proposed method.
Figure 3 shows an overview of the proposed method.
First, continuous cross sections are extracted from the
maternal body of a pregnant woman. Clustering is
performed in each frame, and Grad_CAM
(Ramprasaath. R., 2017) is applied to the clustering
results. The bounding box is generated at the image’s
part with large gradient. The centeral coordinates of
the detected bounding box are obtained with respect
to the continuous cross section. Noise reduction is
performed for the trajectory at that time, and the fetal
birth position in the Uteras is estimated.
3.2 Network
We aim to accurately classify each frame. We utilize
the basic feature extraction layer VGG_16 (Karen S,
2014). As shown in Fig. 4, in order to ensure the
convergence of learning, fine tuning using
ImageNet's weight is performed. We learn the
multilayer perceptron of the full coupling layers of
VGG 16 and its previous convolution layers.
VGG 16 is a convolutional New Jersey network
consisting of a total of 16 layers of 13 convolution
layers and 3 full layers proposed by ILSVRC
(ImageNet Large Scale Visual Recognition
Challenge) in 2014. It has a simple structure which is
not very different from a general convolution neural
net with only a large number of layers. As shown in
Fig. 4, fine tuning is performed from the
layer to
layer of the convolution layers. In the
convolution neural network, shallow layers tend to
extract edges and blobs. On the other hand, deep
layers tend to extract features unique to specific
object that were learnt. In that case, the general-
purpose feature extractor of the shallow layer is fixed
as it is, and the current fetal ultrasound image of the
deep layer weight is relearned. For all binding layers,
four classes are output as "head", "stomach", "leg",
"other". SGD (Stochastic Gradient Descent) is used
for optimization in learning. In the descent method it
is impossible to escape from the problem of local
convergence. However, SGD tends to escape the local
convergence easily due to the influence of random
Figure 4: Model layer (VGG16).
Detecting a Fetus in Ultrasound Images using Grad CAM and Locating the Fetus in the Uterus
183
numbers and it is easy to globally converge (Leon. B,
1991).
Furthermore, our data set includes images of wide
ranges (including head edges and centers) of the head,
abdomen (body) and leg. We do not aim at extracting
some deterministic features, instead, extracting rough
features that likely describes each class.
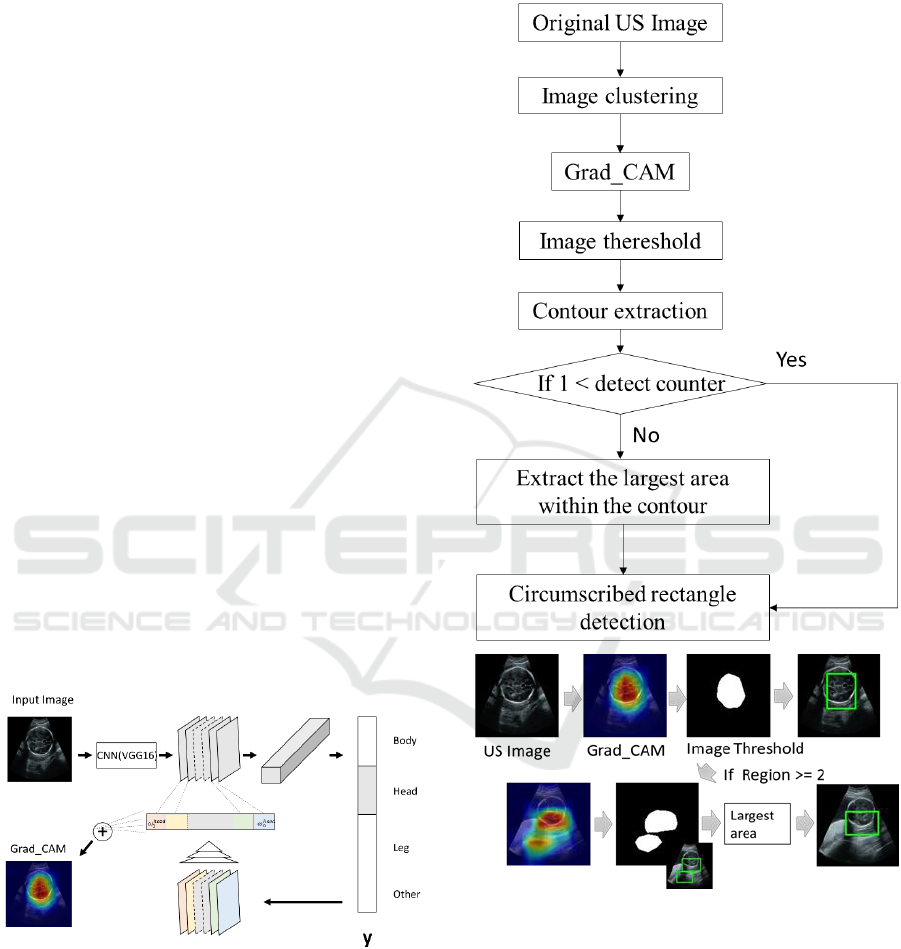
3.3 Grad_CAM
Grad-CAM uses the gradient information flowing
into the last convolutional layer of the CNN to let the
viewer understand the importance of each neuron for
a region of interest. An overview is shown in Fig. 5.
The convolution neural network is roughly divided
into two parts. The first one is a feature extracting part
where the convolution layer and the pooling layer are
stacked in layers, and the second part is an identifying
part which receives the feature output and performs
supervised learning by collating it with the class label.
The identification part usually consists of a multilayer
neural network of all connections, and the final layer
is a soft max layer that converts the feature quantity
to the probability score of each class. The image
points that have a large influence on the probability
score for each class are identified by gradient
averaging. Gradient is a coefficient that indicates the
magnitude of the change that occurs in the probability
score when a tiny change is added to a certain image
part in the feature amount map. The gradient of the
probability score is also large for the image part
where the influence on the class determination is
large.
Figure 5: Conceptual diagram of Grad_CAM.
3.4 Region Localization
By using Grad_CAM, it is possible to visualize which
features in the cross section contribute to judging the
class. Also, since our data set has a wide range in each
class, the visualized features indicate the "likeness"
obtained by the learned network. The area obtained
by this indicator is detected as a bounding box. The
detection of the bounding box is performed according
to Fig. 6.
Figure 6: Detecting bounding boxes.
First, the learned network created in Section 3.2 is
applied to the original ultrasound image. For each
image, a class label (head, body, legs, etc.) is assigned.
After that, the Grad_CAM corresponding to each
label is used. Images’ parts are identified based on
averaging the gradient and the largest probability
score. Meanwhile, gradient equal to or less than the
threshold is removed by Eq (1).
(1)
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
184

where max indicates the maximum value of the
differential coefficient obtained by Grad_CAM for
each image. Note that in Eq. (1), half of the
differential value with respect to the maximum value
is set as the threshold (range). Binarization is
performed using this threshold. In the obtained binary
image, contours are extracted. After then, the
rectangle circumscribing the extracted contour is
obtainnd(Suzuki S, 1985). If the number of the
detected contours (detect counter in Fig. 6) is one, that
contour is detected. If , the
largest detected box is chosen as the final result.
3.5 Object Position Estimation in
Image
The position of the fetus in the image is estimated
using the bounding boxes of each image obtained in
Section 3.4. An example of the image acquisition is
shown in Fig. 7.
Figure 7: Examples of acquiring consecutive frames.
In each image, the bounding box for the detected
object and the central coordinates of the bounding
box are obtained. The central coordinates of the
bounding boxes are obtained for all the continuous
cross sections so that the trajectory of the central point
is obtained. Here, the bounding box is extracted only
when the class labels are head, body, or legs. In case
of “other” label, accurate features are not obtained in
the learning stage. Also, when locating the fetus, the
fetus does not appear in the cross section in many
cases. The bounding box of the "other" class works as
noisy information for locating the fetus; thereby, the
bounding box is not generated.
In addition, the trajectories of the centeral coordinates
of the bounding box detected only by the head, body,
and legs may contain noise due to erroneous detection.
To eliminate this false detection case, noise removed
is performed according to the following procedure.
①Calculate the mean
) of all the images
②Calculate standard deviation (
) of all the images
(2)
③Remove noise outside the range of Eq. (3)
(3)
where a in Eq. (2) and (3) indicate the centeral
coordinates (x, y), n indicates the total number of
frames. The centeral line obtained by this result can
be considerd as the area where the fetus exists.
4 EXPERIMENTS
In this chapter, we present experimental results. First,
the learning results of the network illustrated in
Section 3.2 are explained in Section 4.1. Evaluation
for Section 3.3 is performed in Section 4.2.
Evaluation for Section 3.4 is performed in Section 4.3.
4.1 Classification of Fetal Parts
VGG 16 was used for learning. Fine tuning was
carried out with the total binding layer and the
convolution layer. We prepared 8,000 frames for
training and 2,000 frames for validation. The left and
right movement and scaling were used for the data
augmentation. Batch size is set to 8, and epoch is 30.
The learning rate is set to 0.001. The transition of the
learning result is shown in Fig. 8. The horizontal axis
of Fig. 8 shows epoch. The left vertical axis shows
learning loss and the right vertical axis shows
accuracy
The recall ratio is shown in Table 1. The test data are
combined by 2000 frames (500 frames / class) which
are not used for training.
Figure 8: Relationship between loss and accuracy.
Detecting a Fetus in Ultrasound Images using Grad CAM and Locating the Fetus in the Uterus
185

Table 1: Recall of each part.
Class
Recall
Body
94.0
Head
99.6
Leg
99.8
Other
72.6
High recall ratios were obtained for the head, body
and leg except for “other”. We believe that the other
recall rates are low due to the diversity of the features.
In this study, we estimate the fetal position in the
image,; thereby, we believe that the low recall rate of
the “other” class does not significantly affect the
estimation of feal position.
The confusion matrix of each class is shown in Table
2.
Table 2: Confusion matrix.
body
head
leg
other
body
470
0
25
5
head
0
498
0
2
leg
0
0
499
1
other
34
44
59
363
From Table 2, the “other” class is likely to be
confused with the other classes. It indicates the
uncertainty of the “Other” class. The images of the
“Other” class are easily misclassified by body, head
and leg, which results in a low recall rate.
4.2 Result of Region Localization
An example of the heat map generated by Grad_CAM
is shown in Fig. 9.
The three classes except the “other” class accurately
indicate the part of the fetus. Even from the
classification of the non-experts who are not familiar
with ultrasound images, we could extract the features
of the target part and show them in the form of a heat
map. However, due to the diversity of the “other”
class, the heat map of the “other” class cannot be
easily distinguished. Thus, the VGG-16 network
cannot learn the feature of the “other” class very well.
4.3 Result of Fetal Parts’ Position
Estimation in Images
In this section, we describe the results of applying the
algorithm in Section 3.4. In the experiment, a
phantom (41905-000 US-7 fetal ultrasound
diagnostic phantom "SPACEFAN-ST") of a fetal
Figure 9: Feature extraction using Grad_CAM.
pregnancy medical exercise practice model was used
to fix the condition. In Section 4.3.1, experiments
were conducted on the data acquired as shown in Fig.
7 with the fetal fixed in the center of the image. In
Section 4.3.2, in addition to the image acquired in
4.3.1, the case where the fetal is shifted to the right in
the image and the case where it is shifted to the left in
the image are compared.
4.3.1 A Case in Which the Fetus is Placed in
the Center
In this section, we present the results when we
continuously acquire images from the maternal one
end to the other end while the fetus is at the middle of
the image. Feature extraction of each frame was
performed, and the trajectory of the centeral
coordinates of the generated bounding boxes was
acquired.
Figure 10 shows the results where the horizontal axis
indicates the number of frames, and the vertical axis
indicates the depth for the row direction of the
bounding box. In addition, since the input image to be
learnt is resized to (150, 150), the input image for fetal
part detection is also resized to (150, 150). That is, the
vertical axis of Fig. 9 corresponds to the row of the
image. The number of frames of the continuous cross
section is 761.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
186
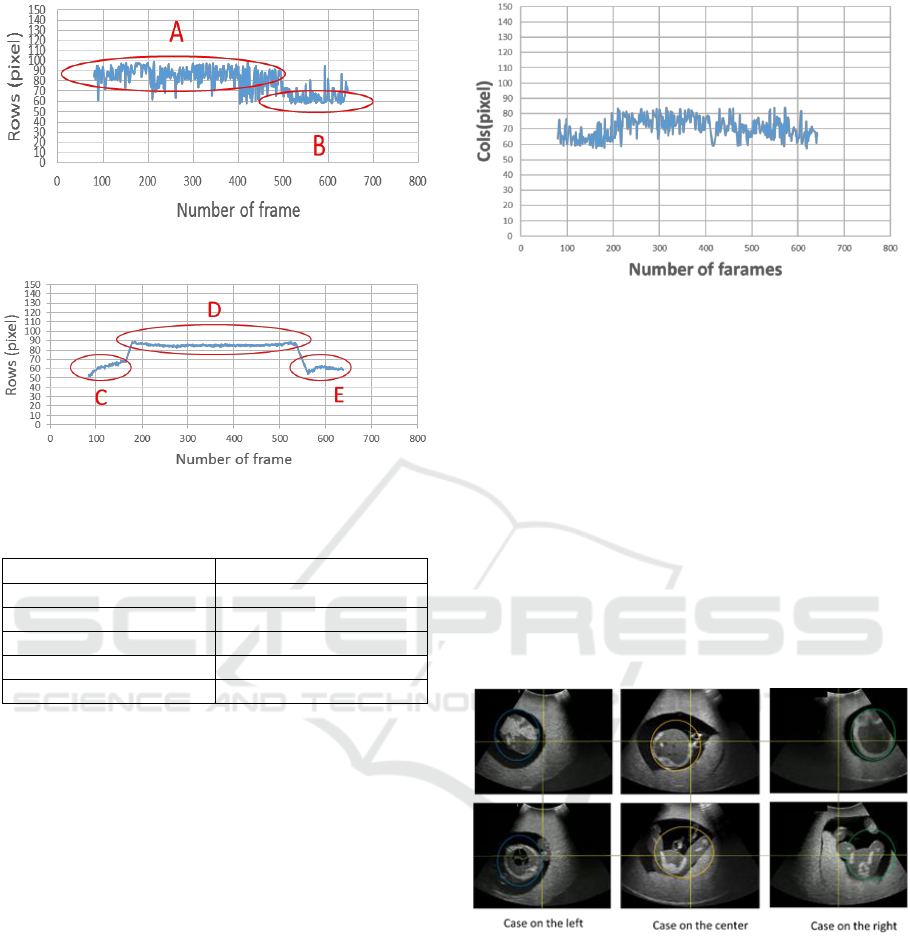
Figure 10: Trajectory of the fetal area(Row).
Figure 11: Shows the Ground truth of Fig. 10.
Table 3: Average of A to E regions in Fig. 10 and 11.
Area
Average depth
A
84.9
B
66.2
C
62.4
D
85.3
E
62.4
Figure 11 shows the ground truth of Fig. 10. The
ground truth is created by the bounding box manually
extracted from the data used in Fig. 10.
Furthermore, Fig. 10 is divided into A and B having
different depths. Figure 11 is similarly divided into C
to E having different depths. The average depths of A
to E are shown in the Table 3. In Fig. 11, C,D and E
correspoud to leg, body, and head, respectivery. In
Fig. 10, the depth of the head region is lost. However,
when comparing A and D, B and E, the errors
between the corresponding areas are very small. On
the other hand, the reason why the head cannot be
detected accurately is that the fetal model "phantom"
has little difference between the head and body, while
the head and body of the real fetus is different.
Next, the result in the left-right direction (cols
direction) is shown in Figure 12.
Figure 12: Trajectory of the fetal area (cols).
As in Fig. 9, the result of Fig. 12 can also show the
center of the bounding box in the images as a
trajectory. From these results, it can be said that it is
possible to estimate how the fetal central axis (fetal
position) is located inside the mother's body.
4.3.2 The Case Where the Fetus is Shifted to
the Left and Right
In this section, when the fetus is shifted to the left and
right of the image in each frame, the trajectory of the
fetal is acquired. The obtained results are compared
with the results shown in Sec. 4.3.1, in which the fetus
is at the “center”. The three kinds of images to be
used are shown in Fig. 13.
Figure 13: Image examples: right, center, left.
As shown in Fig. 13, the images are acquired
continuously. The left column contains 769 frames,
the central column contains 761 frames, and the right
column contains 528 frames. As in Section 4.3.1, the
trajectories of all the three patterns in Fig. 13 are
visualized in Fig. 14.
Detecting a Fetus in Ultrasound Images using Grad CAM and Locating the Fetus in the Uterus
187

Figure 14: Trajectory of fetal areas (comparison of right,
left, center).
If three approximate curves are obtained, it is possible
to visualize the respective positional relationships.
The approximate curve obtained by using the least
squares method for each line is shown in Table 3. The
averages (
) and standard deviations (
) of each
trajectory are also listed in Table 4.
Table 4: Approximate curve parameters.
b
Right
0.0134
90.129
94.362
9.020
Center
0.0042
69.064
70.570
6.524
Left
-0.0011
56.698
56.377
4.145
In Table 4, a is the slope and b is the intercept. At this
time, since the inclination is a small value, it is not
taken into consideration. It can be seen that the
difference between the central trajectory and the
intersection of the left trajectory is narrower than the
difference between the central and the right intercept.
The reason on why the orange and blue lines are close
to each other is due to the fact that the continuous
image acquired so as to pass through the center is
slightly leftward as shown in Fig. 12.
The standard deviation values obtained from the
approximate curves of the loci of "center", "left", and
"right" were 70.57 ± 6.52, 56.38 ± 4.15, 94.36 ± 9.02
respectively. The average indicates the distance
between the fetuses, and the standard deviation
indicates the blur of the central axis of the fetus. The
difference in position appeared in all that, but the
central axis was blurred on the right.
In the future it will be necessary to increase the range
of image augmentation in the left and right direction
and to reduce blurring of the fetal posture. In addition,
it is necessary to improve accuracy by using previous
and next frames.
Let us consider the right case (green), from the results
of the Table 3, the value of σ_a is much larger than
center and left cases, mainly because it is often mis-
classified as “Other” class. 398 frames of all 528
frames are judged as "Other". It is caused by the fact
that many images were missing more than half when
acquiring a continuous cross section. In order to
classify accurately even when it is missing, it is
necessary to increase the amount of movement left
and right during augmentation, and to increase the
number of defect images in a pseudo manner.
However, from the results of Fig. 13 it was suggested
that the trajectory of the fetus could be supplemented
to some extent at present.
4.4 Future Work
The proposed method detects, a fetal region from the
result of Grad_CAM, and a bounding box is
generated, but there is no result that proves it is a
correct answer. It is necessary to show the validity of
the proposed method more quantitatively by having
partnership with a doctor and evaluating accurate
detection by the procedure.
Also, it is necessary to perform comparative
verification of other deep learning frameworks such
as Alex_Net and Res_Net.
In addition, when multiple fetal regions are detected,
the region with the maximal area is outputted. Based
on the results of a few previous frames, the accuracy
of outputting the region could get higher.
We could estimate the position of the fetus in the
image by the proposed method and the fetal position
obtained from the continuous images. Based on these
results, it was difficult to fully determine the standard
cross section for estimating the weight of the fetal in
the past, but it is thought that measurement becomes
easy by detecting the center of the fetal. In addition,
3D ultrasonic machines are also being used more
frequently in recent years.
As a related study, research using 3D data such as
Nambure A. et al.’s research (Nambure A, 2018) on
3D fetal brain’s important feature extraction is
increasing. Furthermore, 3D fetal examination relies
on medical doctors’ experiences more than 2D
examination. The proposed method can obtain the
fetal depth in images; therefore, the estimated results
of the proposed method can be applied to automatic
measure using 3D US machine in the near future.
5 CONCLUSIONS
This paper has proposed an automatic method for
estimating fetal position from ultrasound images
based on classification and detection of different fetal
parts in ultrasound images. Fine tuning is performed
in the ultrasound images to be used for fetal
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
188

examination using CNN, and classification of four
classes is realized. Based on the obtained learning
result and the gradient of the feature obtained by Grad
Cam, a bounding box is generated at the image’s part
with the largest gradient. The trajectory of the center
of the bounding box in each image is obtained as the
position of the fetus. Experimental results are as
follows.
The fetal four class recall ratios were 99.6% for
head, 99.4% for body, 99.8% for legs, 72.6% for legs.
The trajectories obtained from the fetus present in
“left”, “center”, “right” in the images show the above-
mentioned geometrical relationship.
In the estimation of the depth of the fetus,
although the problem remains in estimating the depth
of the head part, the accuracy of estimating the depth
of the fetal body and the leg part is high. In the future,
it is necessary to improve accuracy by using other
deep learning networks and previous and latter
relative frames.
These results indicate that the estimated fetal
position coincides with the actual position very well,
which can be used as the first step for automatic fetal
examination by robotic systems.
ACKNOWLEDGEMENTS
This research is a collaborative study with Ikeda and
Rattan in Iwata laboratory at Waseda University.
Iwata and colleagues are developing TENANG robot
for pregnant women's ultrasound examination.
REFERENCES
Christian F, Konstantinos K, Jacqueline M, Tara P, et. al.
2017, Sononet: real-time detection and localisation of
fetal standard scan planes in freehand ultrasound. IEEE
Transactions on Medical Imaging, 36(11):2204{2215.
Gull I, Fait G, Har-Toov J, et al. 2002, Prediction of Fetal
Weight by Ultrasound: the Contribution of Additional
Examiners. Ultrasound in Obstetrics and Gynecology,
vol. 20, no. 1, pp. 57-60.
Hadlock F. P, Deter R.L, Harrist R.B, Park S.K. 1983,
Computer Assisted Analysis of Fetal Age in the Third
Trimester Using Multiple Fetal Growth Parameters. J
Clin Ultrasound, vol. 11, pp. 313-316.
Hadlock F. P, Deter R., Carpenter R, et.al. 1981, Estimating
Fetal Age: Effect of Head Shape on BPD, American
Journal of Roentgenology. vol. 137, no. 1, pp. 83-85.
Karen S and Andrew Z. 2014, Very deep convolutional
networks for large-scale image recognition. In ICLR,
International Conference on Learning Representations
2015.
Leon. B.199, Stochastic gradient learning in neural
networks. In Proceedings of Neuro-Nˆımes. EC2.
Namburete A, Xie W, Yaqub M, Zisserman A. 2018, Fully-
automated alignment of 3D fetal brain ultrasound to a
canonical reference space using multi-task learning.
Med Image Analysis, vol46, pp.1-14.
Nicolas T, Bishesh K. et al, 2018, Weakly Supervised
Localisation for Fetal Ultrasound Images, 4th
International Workshop, DLMIA 2018, and 8th
International Workshop, ML-CDS 2018, pp.192-200.
Oquab, M., Bottou L, Laptev, I,(2014) Learning and
transferring mid-level image representations using
convolutional neural networks, Proc. CVPR, pp. 1717-
1724.
Razavian, A., Azizpour, H. and Sullivan, 2014, J.: CNN
features off-the-shelf: An astounding baseline for
recognition, Proc. CVPR, pp. 512-519.
Shinzi. T. 2014, Sanka Chouonpa Kensa (Obstetric
ultrasound examination). Japan: Igaku shyoin, p.58.
Suzuki, S, Abe, K., 1985. Topological Structural Analysis
of Digitized Binary Images by Border Following.
CVGIP 30 1, pp 32-46.
Ramprasaath R. Selvaraju, Michael C, et al. 2017, Grad-
CAM: Visual Explanations from Deep Networks via
Gradient-based Localization. IEEE International
Conference on Computer Vision (ICCV), pp. 618-626.
Yuanwei Li, Bishesh K, Benjamin H, 2018, Standard Plane
Detection in 3D Fetal Ultrasound Using an Iterative
Transformation Network. MICCAI 2018: Medical
Image Computing and Computer Assisted Intervention
– MICCAI, vol11070, pp 392-400.
Detecting a Fetus in Ultrasound Images using Grad CAM and Locating the Fetus in the Uterus
189
