
Model Driven Data Management in Healthcare
David Milward
Department of Computer Science, Oxford University, Parks Road, Oxford, U.K.
Keywords:
ISO11179, Metadata, Model Driven Engineering, UML.
Abstract:
Healthcare research depends on the availability of data that is of high quality, that is easy to query, consistent
and current. Traditionally, healthcare data has relied on multiple diverse datasets being integrated by do-
main experts. These integration processes are executed with a high degree of human involvement, integrating
datasets can be time-consuming and can result in the introduction of errors into the data. This paper describes
work to build an integration toolset for healthcare datasets based on the ISO11179 Standard for metadata reg-
istries. It describes issues encountered whilst implementing the standard and shows how these short-comings
were overcome by using techniques from the field of Model Driven Engineering (MDE).
1 INTRODUCTION
A fundamental problem in the UK and elsewhere
is how to make clean data from heterogeneous
healthcare datasets available to researchers easily
and quickly. Much of this information is from re-
search sources such as clinical trials and electronic
patient records, however currently researchers can
spend more time wrangling and cleaning the data
than is spent in analysis, some reports put this at be-
tween 60-80% of time taken in analysis tasks (Press,
2016). Analysis generally involves python and R
scripts which are unique to the researcher that wrote
them, if new information is not in the form that the re-
searcher anticipated then the text has to be re-written,
and checked for accuracy. In an ideal world data
would be input to a data warehouse in a form that
allows the same query to be run repeatedly, because
the data that comes into the repository is guaranteed
to be in the same format, and if the format changes
the dataset and query can be updated to take account
of this without a lengthy script re-write.
One approach to tackling this problem is the use of
the standardized dataset, the idea being is that a set of
data items are defined in the standard, and all report-
ing of any such data-items is made to confirm to the
standard. This will enforce some simple rules such
as a patient identifier in the NHS needs to be an inte-
ger of a certain length, conforming to a certain set of
rules, very often encoded with a regular expression.
This approach helps enormously, but its application
hasn’t been entirely successful to date for several rea-
sons. Firstly, it is impossible currently to mandate
that everyone uses the same standard, or set of stan-
dards. Secondly, some standards are strong in some
areas and weak in others. Thirdly standards evolve.
There are currently a number of different data
standards in healthcare, each one having emerged
from a different specialist area, such as pathology
or pharmaceutical research. If data from heteroge-
neous datasets are presented in one, and only one
of these standards, but for instance, in different for-
mats, some being in XML, some in CSV and some in
RDF, then they can relatively easily be fused with data
from other datasets conforming to the same standard.
Where data standards have clinically endorsed map-
pings between them, then data from different datasets
can easily be merged. Where data is available in
datasets which do not comply with a common stan-
dard, then a set of mappings needs to be made to
merge that data. This is normally carried out using
standard Extract, Load and Transform (ETL) tech-
niques.
Some dataset standards, such as the OMOP CDM
(OHDSI, 2018) claim the title of Common Data
Model (CDM), and aim to be the only dataset def-
inition for the whole industry. In addition, there
are many datasets that have been built up within re-
search organisations or within particular clinical spe-
cialist areas that are in use throughout the U.K.’s Na-
tional Health Service (NHS). Certain of these clinical
datasets were used in this investigation, in particular
COSD, FHIR, SNOMED CT and the NHS Data Dic-
tionary.
Milward, D.
Model Driven Data Management in Healthcare.
DOI: 10.5220/0007391101050116
In Proceedings of the 7th International Conference on Model-Driven Engineering and Software Development (MODELSWARD 2019), pages 105-116
ISBN: 978-989-758-358-2
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
105

This research started out to discover better ways of
integrating datasets, in particular if automated tech-
niques using metadata could be applied to manag-
ing, wrangling and cleaning data used in clinical data
analytics. Some work had already gone into ap-
plying the ideas in ISO/IEC 11179, the ISO stan-
dard for metadata registries, and so initially this was
adopted as a way forward in the research program.
We built and tested the interchange of several datasets
using an ISO11179 compliant metadata registry with
mixed results, we examined problems which arose,
and then built a revised metadata registry built on
model driven engineering principles. Repeating the
same experiments we found that the improved meta-
data registry performed more efficiently, and was eas-
ier for clinicians and healthcare business analysts to
use. We achieved the main research goals of identify-
ing automated techniques for using metadata to man-
age, wrangle and clean dataset far more efficiently
than the techniques previously being used. We then
looked at the problems we had with the ISO11179
standard, and examined what improvements can be
made to the ISO11179 standard to make it more work-
able and effective in achieving the purposes stated in
ISO/IEC11179-1:2015E.
This paper is split into several sections; the section
on related work details previous research efforts to
achieve interoperability between heterogeneous clini-
cal datasets ISO11179 and Model Driven Engineering
techniques. In the section called Background the na-
ture of the problem is described in detail, including
a short summary of some of the main dataset stan-
dards encountered in the course of this research. In
the section titled ISO11179: ISO Standard for Meta-
data Registries the ISO11179 approach to interop-
erability is examined. The next section Evaluation
evaluates the effectiveness in applying ISO11179 to
clinical dataset management. A review of the re-
sults is given in the next section Results, firstly of the
overall research effort, and secondly on the role of
ISO11179. Lastly, there is a section outlining Con-
clusions and suggesting future work. The main con-
tributions of this research are as follows: first, pro-
viding a set of techniques for automating the man-
agement of datasets using metadata, and more specifi-
cally using tools built around a metadata registry, sec-
ond, providing a record of experiences in applying
the ISO11179 to medical dataset management; third,
identifying shortcomings in the ISO11179 metadata
registries standard; fourth, identifying ways to over-
come these shortcomings using model driven engi-
neering principles, and last, the design of an improved
metamodel for healthcare metadata registries.
2 RELATED WORK
The work described in this paper has been informed
by work carried out by colleagues at the University
of Oxford on the CancerGrid project (Davies et al.,
2014), where an ISO/IEC 11179-compliant metadata
registry was developed as detailed in (Davies et al.,
2015). Initially the test software for these studies was
developed using the eXist XML database, but it was
found to have problems scaling once the number of
data elements increased over about 10,000, and so
new work was carried out to build a more scalable
metadata registry using java-based web frameworks.
One of the earliest efforts to apply the princi-
ples of ISO11179 in practice was the caBIG initia-
tive by the National Cancer Institute in the USA,
(Kunz et al., 2009); they built a software develop-
ment kit which allows developers to build web ser-
vice stubs around data elements, (Komatsoulis et al.,
2008), however it doesn’t appear to have been widely
adopted. Indeed there are very few examples of
ISO11179 metadata registries in practice, one study
has used semantic web technology to integrate meta-
data registries, Sinaci and Erturkmen (Sinaci and Er-
turkmen, 2013) describe a semantic metadata registry
framework where Common Data Elements (CDEs)
are exposed as Linked Open Data resources. CDEs
are described in the Resource Description Framework
(RDF), and can be queried and interlinked with CDEs
in other registries using the W3C Simple Knowledge
Organization System (SKOS). An ISO11179 ontol-
ogy has been defined as part of the framework, and
the Semantic MDR has been implemented using the
Jena framework.
Metadata Registries, such as those conforming to
the ISO11179 standard, can help to solve the problem
of data incompatibility, provenance and compliance,
as is indicated in studies such as those conducted by
Ulrich et al. (Ulrich et al., 2016). In this study a hy-
brid architecture consisting of an ISO 11179-3 con-
formant MDR server application for interactively an-
notating and mediating data elements and the transla-
tion of these data elements into Fast Health Interop-
erabililty Resources (FHIR) (HL7-FHIR-Foundation,
2017) resources was used to manage data for the
North German Tumor Bank of Colorectal Cancer.
Tao et al. (Tao et al., 2011) present case studies in
representing HL7 Detailed Clinical Models (DCMs)
and the ISO11179 model in the Web Ontology Lan-
guage (OWL); a combination of UML diagrams and
Excel spreadsheets were used to extract the meta-
models for fourteen HL7 DCM constructs. A criti-
cal limitation of this approach is that the transforma-
tion from metamodels to their ontological representa-
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
106

tion in OWL is based on a manual encoding. Leroux
et al. (Leroux et al., 2012) use existing ontologies to
enrich OpenClinica forms.
Model driven engineering techniques have also
been applied to healthcare problems, Schlieter
et al. (Schlieter et al., 2015) record their experience
gained from using model-driven engineering to im-
plement an application for path-based stroke care;
amongst the lessons learned they recommend using
existing ontological models where possible, and being
prepared to reconcile a heterogeneity of models from
the various stakeholders under a common metamodel.
Atanasovski(Atanasovski et al., 2018) presents a for-
mal meta-model used to specify healthcare process
management in an electronic health record system us-
ing FHIR and OpenEHR. Marcos et al.(Marcos et al.,
2013) describe the implementation of an OpenEHR
system to enable interoperability in clinical trial data
using OpenEHR archetypes. Archetypes are part of a
domain specific language (DSL) which in turn is part
of the OpenEHR standard, discussed in more detail in
the next section, but are described in detail in (Costa
et al., 2011) The problems with integrating data en-
coded using different datasets and terminologies are
clearly identified by Jian (Jian et al., 2007), and so-
lutions using OpenEHR technology are put forward
in (Mart
˜
Anez-Costa et al., 2010).
In the Model Driven Health Tools
(MDHT) (Open-Health-Tools, 2008) project, the
HL7 Clinical Document Architecture (CDA) stan-
dard (Dolin et al., 2006) for managing patient records
is implemented using Eclipse UML tools (Eclipse-
Foundation, 2018). MDHT supports only the CDA
standard, whereas the Model Catalogue can interop-
erate with any metadata standard. The CDA standards
are large and complex: Scott and Worden (Scott and
Worden, 2012) advocate a model-driven approach
to simplify the HL7 CDA, supported by three case
studies: the NHS England ‘InteroperabilityToolkit’,
a simplification of US CDA documents, and the
Common Assessment Framework project for health
and care providers in England.
3 BACKGROUND
As mentioned earlier one goal of this work was to
discover more efficient ways of integrating healthcare
data from heterogenous datasets. During the course of
this work a number of standardised datasets were en-
countered, some being local standards, some National
(such as the Cancer and Outcomes Dataset, managed
by NHS England) and some International (such as
SNOMED CT and OpenEHR).
The challenge here is to build a metadata registry
that can easily store data specifications from all these
varying datasets and standards, to store and make this
information available for other applications which are
able to transform the data from one dataset to another.
This section is designed to give the reader a brief in-
sight into the variability of these datasets.
These healthcare data standards have evolved over
the last 30 years, mostly starting in particular commu-
nities focusing on specific healthcare sub-domains,
such as pathology or laboratory testing. Some evolved
to address local problems, such as defining standard
formats to send laboratory results back to clinicians,
and some started with the broader goals of unifying
global healthcare records with one common standard.
Each dataset has a slightly different way of looking
at data, is used in a slightly different way reflecting
the different sub-domains and cultures that the dataset
evolved from. For instance LOINC is widely used in
laboratories carrying out clinical testing. SNOMED
CT aims at being a clinical terminology which has
terms to be used in the whole of the healthcare do-
mains, however in the sub-domain of laboratory test-
ing LOINC has more granularity, and so there is a
need for mapping between the two data standards.
The data standards that are used in the healthcare
domain, having evolved from specific sub-domains,
tend to use the language adopted by that sub-domain,
and very often the rules of how meaningful sentences
are constructed in the area of laboratory testing for
instance, are not exactly the same rules as are used
in general English. So while these areas look to pro-
vide terminologies, the use of these terminologies can
differ from standard to standard.
In some standards, such as SNOMED CT terms
are designated as pre and post co-ordinated. Pre-
coordinated terms are terms that have compound
terms combined in advance to arrive at a specific des-
ignation, with a specific identifier.
This is illustrated in Figure 1 using the normative
example, taken from the ITSDSO website, showing
the two different ways of expressing a fractured tibia.
The pre-coordinated term, ID-31978002, expresses
the idea of a ”fractured tibia”, whereas the post-
coordinated terms of tibia, ID-12611008 and frac-
ture, ID-125605004 are coordinated during the actual
diagnosis, to form the resultant concept ”fractured
tibia” using the set of SNOMED-CT post-coordinated
terms.
SNOMED-CT is at core maintained as a formal
ontology using the Web Ontology Language (OWL),
and thus the coordination can be carried out using de-
scription logic, as illustrated in (Stevens and Sattler,
2013), and (Rector and Iannone, 2012).
Model Driven Data Management in Healthcare
107
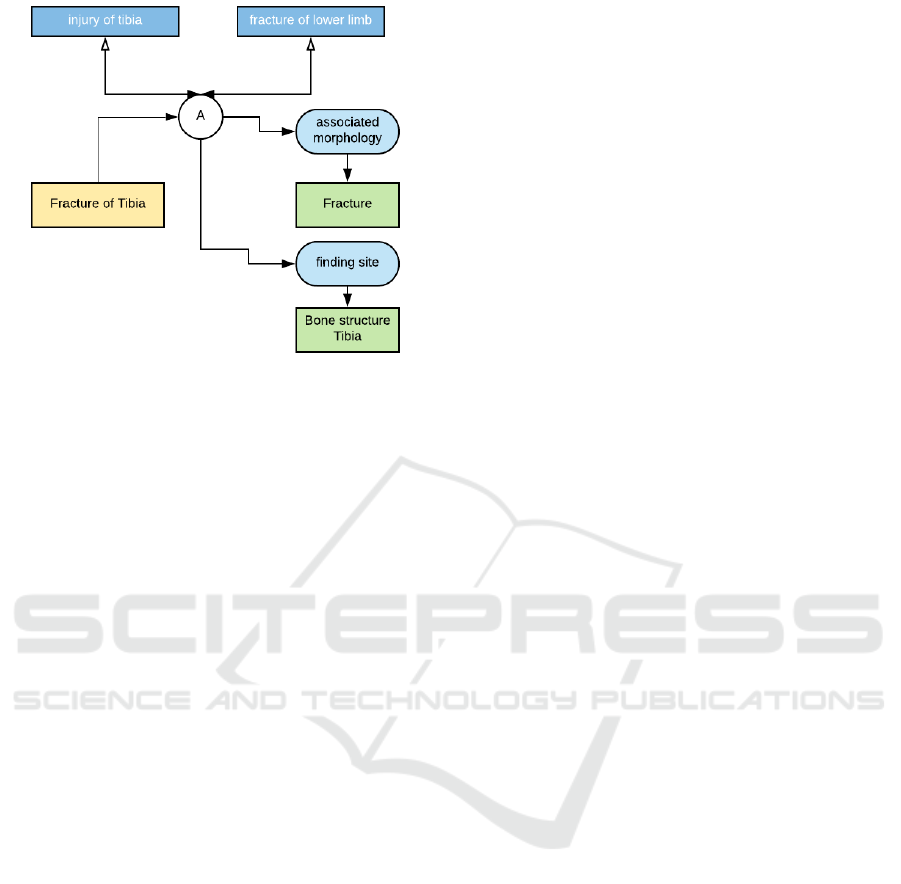
Figure 1: An Illustration of Pre- and Post-Coordinated
Terms in SNOMED CT.
In most healthcare centres today, it is much more
likely to be carried out by other patient record and
clinical diagnosis tools as appropriate, and a mapping
made from the tools’ own dataset to the SNOMED-
CT standard. This throws light on another issue
faced in the process of data collection from hetero-
geneous sources: that the data very often needs to
be transformed, normally using standard ETL pro-
cesses. Datasets that use post-coordinated terms lead
to less flexibility in such transformations, since the
terms used (in this case the post-coordinated Frac-
ture of Tibia as opposed to the pre-coordinated Frac-
ture and Bone Structure Tibia) may not all have di-
rect analogues in the dataset definitions of the data
sources. It is more likely, though not guaranteed, that
a better partial mapping can be made from the pre-
coordinated terms since there are going to be more of
them than from the post-coordinated terms.
3.1 Healthcare Standard Datasets
The data standards being used in healthcare originate
from different specialist areas, use different formats
and have many overlaps, resulting in a number of dif-
ferent viewpoints over which standards are more or
less useful. This section gives an overview and brief
review of some of the main standards encountered,
key considerations from a modelling point of view are
hierarchy depth, granularity, restraints and constraints
and data element relationships. Whilst the domain of
Healthcare might appear to be a single domain to the
outsider, it is split into a number of sub-domains, who
in turn have evolved different process, different ap-
plications and even different uses of language to de-
scribe the work being carried out in the subdomain.
These factors are reflected in the differences between
different datasets, and all contribute to making inter-
operability a complex problem to solve.
3.1.1 SNOMED and Semantic Technologies
The systemized nomenclature of medicine
(SNOMED) began life as the systemised nomencla-
ture of pathology (SNOP) in 1965, originated by the
College of Pathologists(CAP) in the US.
At core SNOMED CT is a set of terms, which
are attached to formally defined concepts, and each is
given a code. It enables different medical conditions
to be given a unique reference, which due to the hier-
archical nature of the core ontology allows clinicians
to go into the appropriate amount of detail. SNOMED
CT has a deep hierarchy of terms, and include pre and
post-coordinated terms, as discussed.
3.1.2 HL7 and FHIR
Health Level 7 is a set of International standards per-
taining to information usage and interoperability in
healthcare, produced by Health Level 7 International
and adopted by both the American National Standards
Institute (ANSI) and the International Standards Or-
ganization (ISO). HL7 has been around for almost 30
years, and in essence it is a set of messaging stan-
dards, definitions and resources, including Version
2.x Messaging Standard, Version 3.x Messaging Stan-
dard, Clinical Document Architecture (CDA), Con-
tinuity of Care Document (CCD), Structured Prod-
uct Labelling (SPL), Clinical Context Object Work-
shop (CCOW) and Fast Healthcare Interoperability
Resources (FHIR).
3.1.3 Read Codes
Read codes are the standard clinical terminology sys-
tem developed by Dr James Read in the 1980’s for
use in the United Kingdom. There are 2 forms of read
codes, a 4-byte version (the original version), a 5-byte
version 2, and version 3 also called Clinical Terms
3 or CTV3. This latter vocabulary was merged with
the US origin SNOMED-RT to form SNOMED-CT.
CTV3 was mandated for use across the NHS in 1999,
it currently consists of a vocabulary of about 200,00
data terms, together with tables that capture various
hierarchies of elements.
3.1.4 International Classification of Diseases
(ICD)
The International Classification of Diseases(ICD) is
managed by the World Health Organization (WHO),
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
108

the authority for health matters in the United Na-
tions.[2] The ICD is a health care classification sys-
tem, it’s primary purpose is to provide a set of di-
agnostic codes for diseases and disease classification.
This includes nuanced classifications of a wide vari-
ety of items such as signs, symptoms, abnormal find-
ings, complaints, social circumstances, and external
causes of injury or disease.
3.1.5 OpenEHR - ISO EN 13606
OpenEHR is an open standard for electronic health
records, which is defined by the international standard
ISO13606 (ISOTC215, 2008). From the perspective
of data management the standard provides a reference
model and a number of archetypes, which describe
datasets from a clinical perspective and templates,
which provide instantiations of those archetypes for
specific use cases. The standard covers not only an
architecture definition, but also a domain specific lan-
guage for storing, managing and querying healthcare
records.
3.1.6 OMOP CDM
The Observational Medical Outcomes Partnership
(OMOP) Common Data Model(CDM) came out of
work started in 2008 by a public-private partnership
in the USA, between the FDA and the pharmaceutical
industry to research the utility of using existing health
care databases to evaluate safety issues relating to cur-
rently approved and available drugs.
3.1.7 LOINC
The Logical Observation Identifiers Names and
Codes (LOINC), was started in 1994 with goal of
codifying laboratory specimens and tests in a clinical
context. It is designed as a data standard for labora-
tory tests and results, anything that can be observed
or tested in relation to a patient’s healthcare. Part
of LOINC can be used for normal observations, and
part of it for laboratory results. It is at core a large
terminology, more detailed in many areas of pathol-
ogy than SNOMED CT. It is very often used in con-
junction with both SNOMED CT and HL7, so that
messages sent requested laboratory tests may be cod-
ified in HL7 using LOINC, and responses may use
SNOMED in addition to LOINC.
3.1.8 NHS Data Dictionary
The NHS Data Dictionary is maintained by NHS Dig-
ital as a UML model, however it is publicly avail-
able as either HTML files, viewable on a website, or
as XML/XSD files which can be downloaded. The
model is relatively straightforward, and is used for
most of the reporting carried out by NHS Digital in
the UK.
4 ISO11179: ISO STANDARD FOR
METADATA REGISTRIES
Metadata can be recorded using a metadata registry;
a metadata registry (MDR) is a toolkit which allows
definitions of datasets to be stored, curated and man-
aged. Metadata is usually defined as data about data;
this is an unfortunate definition, in that it can be inter-
preted in a variety of ways. Our work relates to data
management and for most data management purposes
it is normally taken to mean data which defines the
structure of data, although data about the governance,
provenance and other aspects of that data can also be
relevant, and thus included in a metadata registry. By
storing the definitions of every data element and also
the relationships between data elements in a metadata
registry a map of all the data elements and data flows
in an organization or domain can be created. Such a
map can be used to manage, understand and curate
the datasets being used. The ISO standard 11179 is a
standard for metadata registries, although this desig-
nation is extended in the body of the standard.
The ISO11179 (ISOJTC1, 2015) standard, was is-
sued in 2015, is composed of 6 parts:
• ISO/IEC 11179-1:2015 Framework (referred to as
ISO/IEC 11179-1)
• ISO/IEC 11179-2:2005 Classification
• ISO/IEC 11179-3:2013 Registry metamodel and
basic attributes
• ISO/IEC 11179-4:2004 Formulation of data defi-
nitions
• ISO/IEC 11179-5:2015 Naming and identification
principles
• ISO/IEC 11179-6:2015 Registration
The standard is comprehensive, however we are only
considering some key aspects of Part 3 in this re-
search. Part 7 called Datasets, is planned and is cur-
rently under review. This introduces a metamodel for
datasets into the standard, however this was not read-
ily available at the time that this research was carried
out, and is not considered here. To quote from the
standard itself:
ISO/IEC 11179 addresses the semantics of
data, the representation of data and the reg-
istration of the descriptions of that data. It is
Model Driven Data Management in Healthcare
109
through these descriptions that an accurate un-
derstanding of the semantics and a useful de-
piction of the data are found.
The purpose of ISO/IEC 11179 is to promote the fol-
lowing:
• standard description of data
• common understanding of data across organiza-
tional elements and between organizations
• re-use and standardization of data over time,
space, and applications
• harmonization and standardization of data within
an organization and across organizations
• management of the components of descriptions of
data
• re-use of the components of descriptions of data
Part 3 of the standard provides a registry meta-
model, specified using UML diagrams, and it was
thus chosen as the starting point for building this im-
plementation. There is a warning at the beginning
of Part 3, stating that this part prescribes a concep-
tual model, not a physical implementation This part
of ISO/IEC 11179 also prescribes a list of basic at-
tributes (see clause 12) for situations where a full con-
ceptual model is not required or not appropriate. The
other 5 parts were used to inform the core metadata
registry metamodel as specified in ISO11179: Part 3.
4.0.1 Part 3 - Registry Metamodel and Basic
Attributes
The objectives of the metadata registry metamodel are
defined, in the standard, as:
• Providing a unified view of the concepts, terms,
value domains and value meanings
• promoting a common understanding of the data
described
• providing the specification at a conceptual level to
facilitate the sharing and reuse of the contents of
the implementations
The standard continues to split the metamodel up into
6 packages, Basic, Registration, Concepts, Binary re-
lations, Data description and Identification, Designa-
tion and Definition.
In section 4 it is pointed out that clauses 7-9 are
needed to implement a Concept Systems Registry,
clause 10 will allow the implementation of an Ex-
tended Concept Systems Registry, and clause 11
specifies a metadata registry, whereas an extended
metadata registry will implement all clauses 7-11.
Our initial scope was to implement an extended meta-
data registry as described in clauses 7-11.
4.0.2 Data Description Package
This package specifies a metamodel for handling
data, and although it references other packages which
are mostly dealing with the more administrative as-
pects of registering metadata, it primarily puts for-
ward a conceptual metamodel for handling data.
Hence for the purposes of data interoperability it is
the most relevant part of the standard.
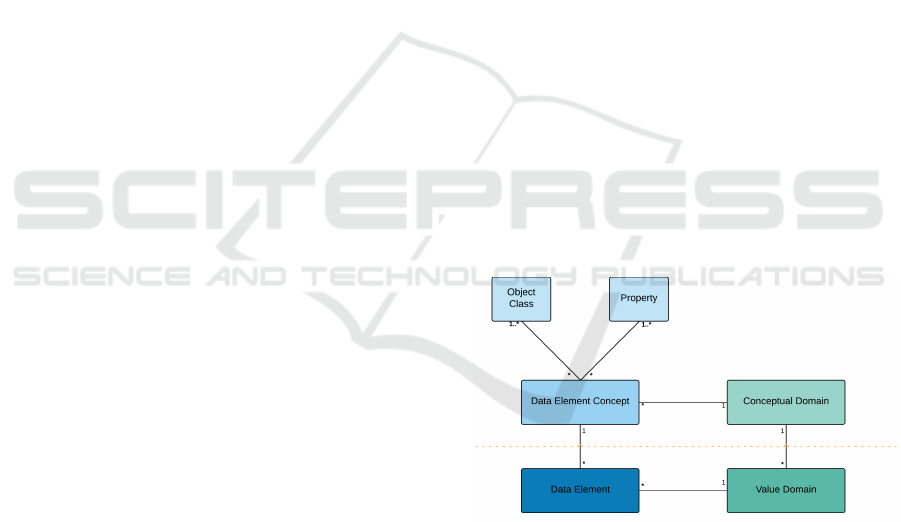
The core model given for data description in
ISO11179 is that reproduced in Figure 2, it shows the
linkage between a Data Element, a Value Domain,
a Conceptual Domain and a Data Element Concept.
The area above the dotted red line is defined as the
semantic or conceptual level, whereas the area below
the red dotted line is defined as the representational
level. The assumption is that the Data Element and
Value Domain are objects which are being registered
and classified, as per the processes defined in other
parts of the standard.
This arrangement can be illustrated by the idea of
a visit to the doctor, we can define a concept called
reason for visit to healthcare centre and call this a
data element concept, and from this we would imple-
ment a data element called reason for attendance and
perhaps represent that with a set of enumerated codes,
each representing a different reason. In ISO11179
structuring we would split the data element concept
into an object class: Person, and a property: Reason
for clinic attendance.
Figure 2: ISO 11179 Data Description.
4.0.3 Evaluation of ISO/IEC11179(2013)Part 3
Part 3 of the standard, titled Registry Metamodel and
basic attributes, and revised in 2013, was taken as the
reference point for building the Metadata Registry im-
plementation. In reviewing the standard several prob-
lems came to light, especially with regard to imple-
mentation, and especially with regard to any kind of
conformity. These are listed as:
• ISO11179 introduces representational items, such
as Conceptual Domain, Data Element, Data El-
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
110

ement Concept, etc, indicating that they are part
of the standard or ideal metadata registry meta-
model with no indication of how the ISO11179-
compliant models so defined are generated, used
or related, nor how data can be transformed into
this particular model or what the actual advantage
is over any other metamodel/model.
• Partial UML models are specified at different lev-
els of granularity, with no explicit connection to
show how they relate.
• The text does not allow an overall model to be
built with any degree of certainty of whether or
not it conforms to the standard.
• Basic types used in the metamodel include types
which in most computer science contexts would
be viewed as derived types, this makes implemen-
tation needlessly difficult and confusing.
• The introduction asserts that metadata registry is
specified in the form of a conceptual data model,
however, despite references to other standards,
and a brief explanation in Appendix E, no defini-
tive explanation of what is meant by conceptual
data model is provided.
• The examples provided are very concept specific,
for instance, the example of country codes works
for concepts which are used as a list, but many
concepts used in healthcare are not used in such a
straightforward fashion.
• Many concepts and terms are described, some are
specified, and some specifications overlap with
other definitions; for instance value domains are
specified with the same definition that is used to
describe data types.
• The UML models provide a great deal of detail
for each sub-section of a metadata registry, how-
ever no system diagram or clear description is pro-
vided, it is therefore impossible to build a working
system based on the UML diagrams alone, con-
siderable interpretation is required, which detracts
from the specification provided.
The standard is declared as being a standard for meta-
data registries, and although it contains a lot of dis-
parate ideas on the subject of interoperability which
can be applied to a metadata registry, no core set of
definitions, core language or metamodel was found
that could be used as a measure for conformance.
Standards by definition should be conformed to, and
whilst conformance is mentioned, it is lacking a clear
set of definitions which can be used as a measure-
ment for conformity. Therefore the goal of building
an ISO11179 conformant metadata registry was aban-
doned early on, when it became apparent that clear
objective conformance criteria were not present in the
standard, despite the subject of conformance being
discussed. That said, there is much in the standard
document which can be usefully incorporated into the
design of a metadata registry, and development con-
tinued with a view to include those aspects of the stan-
dards which could be shown to be beneficial to the
construction of a metadata registry.
5 IMPLEMENTATION
In the initial design work, the UML diagrams con-
tained in Part 3 section 11 was taken to be the basic
metamodel around which the core metadata registry
would be built.
As detailed here, this very quickly became un-
workable, mostly as a result of user’s being unable
to translate data structures into the form dictated by
the ISO11179 metamodel.
5.1 Implementation of ISO11179 UML
Metamodel
Initially work began by implementing the UML mod-
els as specified in the standard (part 3, section 11),
however when the initial prototype was run, many
pieces of information were identified as being loaded
more than once. Due to the model provided, there is
an overlap of the conceptual structure of the meta-
model, and the logical structure of the metamodal, al-
though no such reference is available in the standard
itself. Initially a basic domain model was built, using
a Grails 2.4.3 toolkit, using the following basic repre-
sentational items shown in the first column of Table 2
The work was then shown to analysts and clin-
icians experienced in building healthcare datasets,
with a view to having them enter suitable datasets
and then take part in developing the data set curation
functionality around the ISO11179 conformant meta-
model.
5.2 Concerns over ISO11179
Conformance
The first major problem was in specifying exactly
what metadata should be input into the prototype
metadata registry, in particular how to translate or
transform existing models or meta-models into the set
of constructs defined and discussed in the standard.
To illustrate this issue, consider taking a data item
from an existing medical dataset, in this case COSD,
as shown in Figure 3.
Model Driven Data Management in Healthcare
111
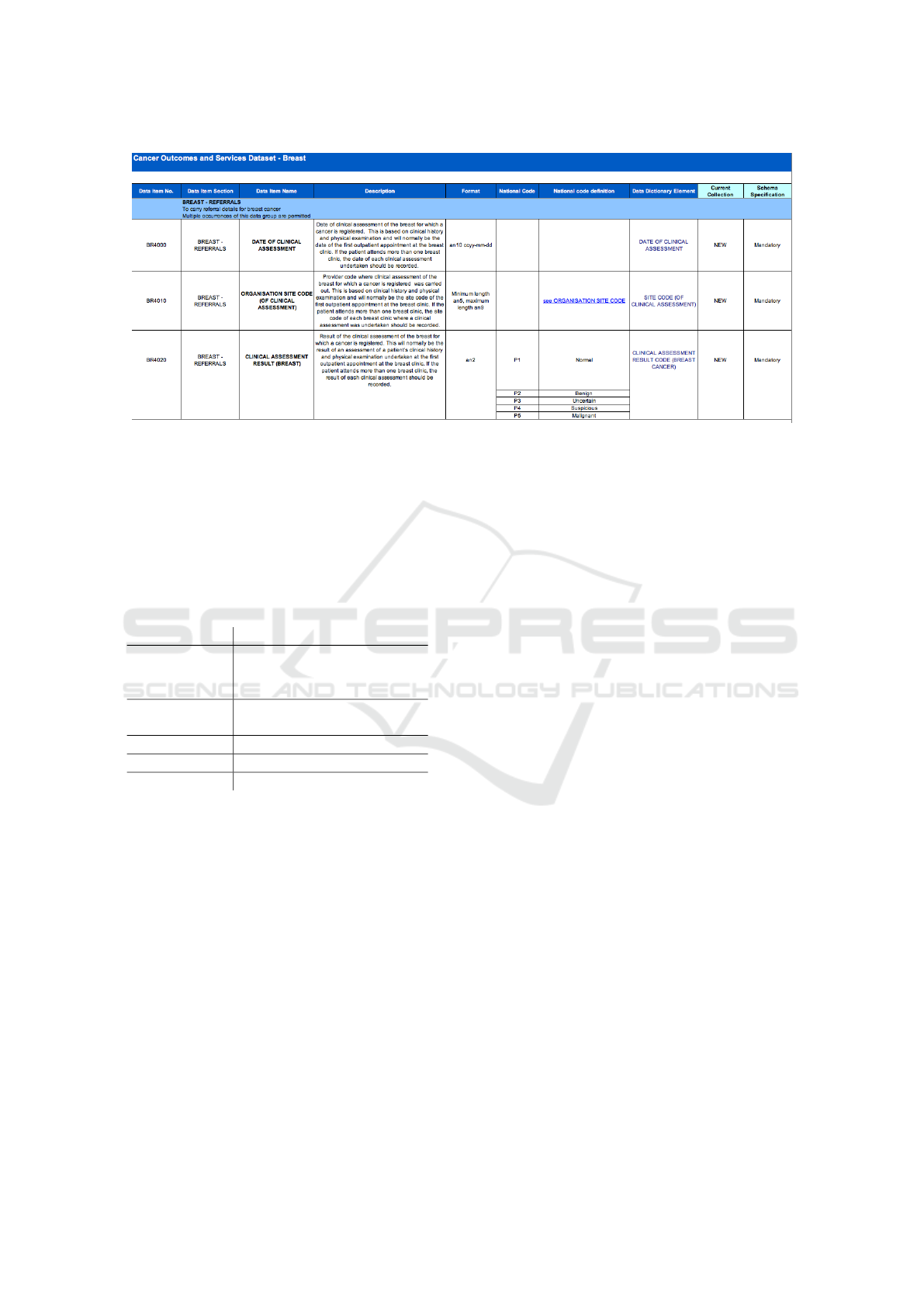
Figure 3: COSD Dataset Excerpt.
Using ISO11179 we take the date of clinical as-
sessment as a data element, however it would be spec-
ified with the patient details, since conceptually one
would need to describe the context of the data item,
this results in a data element which has an object prop-
erty of patient as an integral part of the construct. This
is illustrated in Table 1
Table 1: ISO11179 Cancer Referral Representation.
ISO Artefact Description
Data Element Patient and Date of clinical
Assessment in form ccyy-
mm-dd
Data Element
Concept
Patient and Date of clinical
Assessment
Value Domain Date in form ccyy-mm-dd
Object Class Patient
Property Diagnosis Date
However if one is trying to enter the metadata for
the Date of Clinical Assessment shown in the first row
of Figure 3, there is an immediate disparity in that no
object class is specified, available or immediately ob-
vious from the spreadsheet. There is mention of the
outpatient or patient in the description, however it is
not in the view of the analyst preparing the dataset
of any significance, and therefore was not included
in a separate column. Therefore there is immediately
a dilemma, do we enter outpatient, patient or simply
leave the object property blank? The next issue is the
idea of having the data element, if we drop the Pa-
tient from the name and then call the data element
Date of Clinical Assessment, what then is the Data
Element Concept? and how is it different from the
description of the Data Element? Should the Data El-
ement Concept include the patient? There is no obvi-
ous answer, and from a user perspective it appears that
a simple set of dataset metadata, i.e. column headings
on a spreadsheet, are being transformed into some-
thing more complex in order to manage them, but that
management can only be carried out by experts versed
in ISO/IEC11179.
5.2.1 User Difficulties
In the first few weeks of trying to enter standard ex-
isting healthcare datasets into the prototype metadata
registry many objections were encountered from users
experience with existing healthcare datasets, of the
kind documented in the previous section. Metadata
was seen to be entered twice or three times need-
lessly, the difference between the description of a
Data Element and a Data Element Concept was not
understood. Likewise the difference between a Value
Domain and a Data Type whilst apparent in theory,
was in practice not apparent, since the models being
generated were not implementation specific. There-
fore a representation of a set of numerical values
would in nearly all cases be represented by the same
data type, for instance the date of clinical assess-
ment would have a value domain of date and a data
type of date, which would then be implemented in
a particular system as appropriate, e.g. text string,
org.joda.time.format.DateTimeFormat as appropriate
by the system concerned.
5.3 Update to Meta-model
After a few weeks it was decided to update the meta-
model, at first, the number of ISO11179 elements was
reduced, however this still didn’t gain any traction
with users, who found the system confusing, counter-
intuituve, time-consuming and needing a lot of extra
work to understand the new language constructs in-
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
112
troduced by the standard. As a result the development
went through a two iterations to arrive at the current
model, the initial prototype we refer to as version 0.x,
the second as 1.x and the third as 2.x. The third, has
been fairly successful as is being used at over 7 hos-
pital trusts and healthcare research centres in the UK
currently. The changes in core constructs are shown
in Table 2.
Table 2: Metamodel Domain Constructs.
ISO Artefact (v.0.x) Iteration 2 (v.2.x)
Described Concep-
tual Domain
-
Object Class DataClass
Property -
Data Element Con-
cept
-
Data Element DataElement
DataType DataType
Value Domain -
Described Value Do-
main
-
Enumerated Value
Domain
EnumeratedType
Permissible Value -
Enumerated Concep-
tual Domain
-
Relations Relationship
- RelationshipMetadata
- RelationshipType
Classification -
Concept System
Concept -
Classifiable Item CatalogueElement
Measurement Unit MeasurementUnit
Measure Class -
Dimensionality -
- Asset
- AssetFile
- ExtensionValue
- Mapping
- DataModel
- DataModelPolicy
- PrimitiveType
- ReferenceType
- Tag
- ValidationRule
The first iteration resulted in version 1.x of the
metadata registry, which went into service in Ge-
nomics England in 2015, however it met with many
criticisms from users, and a complete overhaul was
undertaken. This time a different approach was used,
and the basic metamodel redeveloped, informed more
by feedback from data analysts than reliance on the
ISO/IEC11179 standard. The domain metamodel was
developed using XText, which allowed for the fast
iterative development of the metamodel, using the
Eclipse toolkit. Once this was established the domain
model was implemented using the Grails framework
(v2.5.6), which allowed much of the existing code-
base to be re-used.
5.3.1 MDML: A DSL for Metadata
Management
Revising the core metamodel required examining a
wide range of existing healthcare datasets, such as
the ones mentioned in the introduction, and review-
ing the conceptual background, language, context and
purposes to which they were being used. Various
modelling methodologies were examined, however
the ECore/XText framework provided a relatively
straightforward way of testing ideas quickly. A gram-
mar was developed using XText which was called the
Metadata Management Language (MDML), and from
this the most recent metadata registry was developed.
When building this there were a number of fea-
tures or requirements, that the Healthcare professions
we dealt with required. These were absent in the
ISO/IEC11179 standard, and are listed below:
• The ability to group data elements, in particular
handle hierarchical groupings which are so com-
mon in most datasets.
• The ability to manage, compare and map groups
of models as well as individual data elements.
• The ability to uniquely identify particular data el-
ements, groupings, and identify a publishing state,
i.e. is the dataset a draft, a current dataset, a su-
perseded (but still used)dataset.
Some of these issues are touched upon in the stan-
dard, for instance data elements can be grouped us-
ing the construct of object property, however there is
no notion of layers of grouping. With SNOMED CT
there are 13 layers of hierarchy, with links between
layers, and this kind of structure needs to be modelled
and managed with the metadata registry.
The XText domain model included all the con-
structs from column 3 of Table 2, and provided a
grammar which was then used to generate a set of
domain objects, which in turn were able to repre-
sent the various healthcare datasets being used. The
core enabled the automatic generation of grails do-
main classes using the code generation capabilities
of XText. For illustrative purposes we show the key
metadata registry structure in Figure 4.
Model Driven Data Management in Healthcare
113

Figure 4: Core MDR (Ecore) Metamodel.
The structure illustrated here allows the metadata
registry to capture data elements, which is at the core
of the standard. It also allows each element to have
a status, which allows it to conform to the publishing
aspects of the standard. By making each item in the
registry a class inheriting from an abstract Catalogue
Element each data element is captured by a unique
identifier and a status relating to its publishing status.
In the ISO standard metamodel the data element is the
primary artefact under consideration, however there is
no obvious direction on how groups of data elements
should be handled.
In practice data elements appear in groups,
whether spreadsheets, database tables or CSV files.
In the data analysis business, groups of data need to
be brought together and to do this datasets need to
be mapped and transformed. In our first and second
models it was easy to generated lists of data elements,
however they were nearly always grouped using sev-
eral layers of hierarchy into different entities, which
made mapping and transformation difficult.
Further a DataModel is able to add in any num-
ber of extensions, which can be used to augment par-
ticular data elements for instance to mark them as
user interface items so that user interfaces can be au-
tomatically generated. In addition imports can be
added from other DataModels, so that new datasets
can be created using data elements from several dif-
ferent datasets. This metamodel evolved over a period
time in order to solve the problems of integrating data
from a wide variety of heterogeneous sources, it is
closely based around UML/Ecore, but is in essence a
domain specific language or metamodel for data inte-
gration.
5.3.2 Core Elements of MDML
The main constructs of DataClasses, DataElements
and DataTypes are common to most technical spaces,
and can be used to group and manage data elements
in way that is easy for data analysts and developers
to grasp. Metadata can then be attached to any of
these core elements, so that security classifications,
and governance directives, found in other parts of the
ISO11179 standard can easily be added to particular
DataElements or DataClasses.
AT core MDML provides a metamodel, in essence
a language for data structures, which allows addi-
tional metadata to be added on through a simple
mechanism of extension values and tags. It allows
datasets to be grouped and classified in a flexible
and manageable way, so that one registry could hold
several common healthcare metamodels, say FHIR,
OMOP and LOINC, and build local models based on
all three. There is a clear identification system built
into the language which means that each version is
separately identified, so that any mappings between
FHIR and OMPOP for instance will be detailed to
particular versions of the datasets.
There is a clear publishing cycle built into the
metamodel, which allows change for draft models,
but not for finalized models, therefore data elements
in finalized models can be imported into other (draft)
datamodels, but no new elements can be imported in
currently finalized models. This is an important man-
agement feature, derived from the ISO standard which
has been built into the language.
6 RESULTS
By using the metadata registry Genomics England
was able to specify exactly what data they required
from UK Hospital Trusts by building their own data
models centrally online. The other organizations are
then able to use these models to verify that outbound
data is valid, and thus ensuring that data won’t be re-
turned.
The main technique used is to import a new
dataset into the metadata registry, normally in csv or
excel format, to configure a mapping using XML, and
then import the dataset definition into the metadata
registry directly. From this new model a number of
artefacts such as spreadsheets and XML Schema files
could be generated from one source. This eliminated
any doubt arising from spreadsheets which were dis-
seminated after meetings. Instead the meetings could
be held online, and the changes were immediately vis-
ible to all parties. Spreadsheets and XML files could
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
114

then be generated for local form or code generation,
and rules could be attached to data elements and data
types which could be automatically downloaded via a
REST interface, rather than by disseminating spread-
sheets and documents.
The MDML based metadata registry has been in
use for the past 2 years at Genomics England, as well
as at 7 other NHS Hospital Trusts in London. It’s core
use is to manage datasets which are used by Genomics
England to specify dataset requirements, a dataset or
model is defined in the metadata registry, this is then
used to automatically generate output specifications
as excel, XML or as Case Report Forms. The latter is
a specification for generating forms in the Open Clin-
ica system, but which can automatically be loaded
into Open Clinica to generate both the forms and data
repository to store the data. Rules, embedded into the
DataTypes as regular expressions, or as groovy code,
are then used to validate datasets.
Data curation, which previously was carried out
using excel spreadsheets, and involved regular phys-
ical meetings between data managers and clinical
leads from all over the UK now takes place online.
The models which are generated are given a unique
identifier, and once all parties agree to issue a new
model it is finalized thus preventing further change
to that version of the model. At any point in time
the model, and it’s history can be referred to online,
removing doubt and ambiguity over what the con-
stituents are, further the model can be accessed using
a REST interface, making it available for code gener-
ation utilities remotely. The results of using the meta-
data registry and toolkit are listed below:
• Reduction in time and effort in creating and cu-
rating datasets over spreadsheets and document-
based techniques.
• Ability to clearly access the current version of a
dataset and verify that it is the correct version, us-
ing unique identifiers.
• Ability to generate artefacts based on the data-
model, such as forms, XML and XML Schemas
automatically.
• Ability to validate data against the model au-
tomatically using rules stored against individual
data elements.
• Ability to map between different datasets in a de-
tailed and precise fashion, again with the use of
unique identifiers.
7 DISCUSSION
By applying MDE principles a metadata registry was
built that satisfied the requirements of this research
organisation for managing heterogeneous dataset. At
the beginning of the work, much time and effort was
put into implementing the international standard for
metadata registries which is either lacking in clar-
ity, or un-informed by the healthcare use cases that
were encountered in this research effort. Whilst much
in the standard was informative and relevant to the
more general issue of data integration, the clear lack
of a consistent metamodel or language around which
to build a metadata registry was a great disappoint-
ment. Metadata registries are becoming common-
place in enterprise architecture today, especially since
automatic management of large datasets, big data, is
extremely difficult without them. So it was very dis-
appointing that applying the ISO/IEC11179 standard
proved so difficult in this instance. It must be pointed
out that the shortcomings found and described in this
paper only relate to 2 or 3 clauses in a standard which
is several hundred pages in length, however they did
cause significant problems in our development effort
and it is hoped that clarifications and improvements
to the standard will be made in due course.
8 CONCLUSION
There were a number of problems which arose from
attempting to build a metadata registry which com-
plies with the ISO/IEC11179 standard in the early
stages, these have been documented, and steps taken
to correct the problems encountered. In the course
of correcting these problems a model driven engi-
neering approach was taken, and an effective set of
tools were built based around a revised metamodel
for metadata management. The open source toolkit
(MetadataWorks, 2018) developed from this research,
has proved very effective, being used to simplify and
speed up the work of data curation, data-wrangling
and data-cleaning. In addition an MDE-based toolkit
was built which provided a REST interface and al-
lowed third parties to carry out automated data vali-
dation. Future work on extending the model is antic-
ipated, in particular to produce automated tool chains
which can generate both the input forms, repositories
and mapping capabilities for heterogeneous datasets.
Model Driven Data Management in Healthcare
115

ACKNOWLEDGEMENTS
I would like to acknowledge the help of Adam Mil-
ward, Kathy Farndon, Amanda O’Neill and Samuel
Hubble at Genomics England, and Jim Davies,
Charles Crichton, Steve Harris and James Welch at
the University of Oxford.
REFERENCES
Atanasovski, B., Bogdanovic, M., Velinov, G., Stoimenov,
L., Dimovski, A. S., Koteska, B., Jankovic, D.,
Skrceska, I., Kon-Popovska, M., and Jakimovski, B.
(2018). On defining a model driven architecture for
an enterprise e-health system. Enterprise Information
Systems, 12(8-9):915–941.
Costa, C. M., Men
˜
A¡rguez-Tortos, M., and Fern
˜
A¡ndez-
Breis, J. T. (2011). Clinical data interoperability based
on archetype transformation. Journal of Biomedical
Informatics, 44(5):869 – 880.
Davies, J., Gibbons, J., Harris, S., and Crichton, C. (2014).
The cancergrid experience: Metadata-based model-
driven engineering for clinical trials. Science of Com-
puter Programming, 89:126–143.
Davies, J., Gibbons, J., Milward, A., Milward, D., Shah, S.,
Solanki, M., and Welch, J. (2015). Domain-specific
modelling for clinical research. In SPLASH Workshop
on Domain-Specific Modelling.
Dolin, R. H., Alschuler, L., Boyer, S., Beebe, C., Behlen,
F. M., Biron, P. V., and Shvo, A. S. (2006). HL7 Clin-
ical Document Architecture, release 2. Journal of the
American Medical Informatics Association, 13(1):30–
39.
Eclipse-Foundation (2018). Eclipse mdt uml2 tools.
https://eclipse.org/modeling/mdt?project=uml2//.
HL7-FHIR-Foundation (2017). Fast health interoperability
resources. https://www.hl7.org/fhir//.
ISOJTC1 (2015). Io11179 international stan-
dard for metadata registries. http://metadata-
standards.org/11179//.
ISOTC215 (2008). Health informatics – electronic health
record (ehr) standard. http://www.en13606.org/.
Jian, W.-S., Hsu, C.-Y., Hao, T.-H., Wen, H.-C., Hsu, M.-
H., Lee, Y.-L., Li, Y.-C., and Chang, P. (2007). Build-
ing a portable data and information interoperability
infrastructure framework for a standard taiwan elec-
tronic medical record template. Computer Methods
and Programs in Biomedicine, 88(2):102 – 111.
Komatsoulis, G. A., Warzel, D. B., Hartel, F. W., Shanbhag,
K., Chilukuri, R., Fragoso, G., de Coronado, S.,
Reeves, D. M., Hadfield, J. B., Ludet, C., et al.
(2008). caCORE version 3: Implementation of a
model driven, service-oriented architecture for seman-
tic interoperability. Journal of Biomedical Informat-
ics, 41(1):106–123.
Kunz, I., Lin, M.-C., and Frey, L. (2009). Metadata
mapping and reuse in caBIG. BMC Bioinformatics,
10(Suppl 2):S4.
Leroux, H., McBride, S., Lefort, L., Kemp, M., and Gib-
son, S. (2012). A method for the semantic enrich-
ment of clinical trial data. In Health Informatics:
Building a Healthcare Future Through Trusted In-
formation; Selected Papers from the 20th Australian
National Health Informatics Conference (HIC 2012),
volume 178, page 111. IOS Press.
Marcos, M., Maldonado, J. A., Mart
˜
Anez-Salvador, B.,
Bosc
˜
A¡, D., and Robles, M. (2013). Interoperabil-
ity of clinical decision-support systems and electronic
health records using archetypes: A case study in clini-
cal trial eligibility. Journal of Biomedical Informatics,
46(4):676 – 689.
Mart
˜
Anez-Costa, C., Men
˜
A¡rguez-Tortosa, M., and
Fern
˜
A¡ndez-Breis, J. T. (2010). An approach for
the semantic interoperability of iso en 13606 and
openehr archetypes. Journal of Biomedical Informat-
ics, 43(5):736 – 746.
MetadataWorks (2018). Metadata exchange (open source)
toolkit. https://github.com/MetadataConsulting/-
ModelCataloguePlugin//.
OHDSI (2018). Ohdsi common data model. https://www.
ohdsi.org/data-standardization/the-common-data-
model//.
Open-Health-Tools (2008). Model driven health tools.
https://projects.eclipse.org/proposals/model-driven-
health-tools.
Press, G. (2016). Cleaning big data: Most-time-consuming,
least enjoyable data science task. Forbes.
Rector, A. and Iannone, L. (2012). Lexically suggest, logi-
cally define: Quality assurance of the use of qualifiers
and expected results of post-coordination in snomed
ct. Journal of Biomedical Informatics, 45(2):199 –
209.
Schlieter, H., Burwitz, M., Sch
¨
onherr, O., and Benedict, M.
(2015). Towards model driven architecture in health
care information system development. In 12th In-
ternational Conference on Wirtschaftsinformatik (WI
2015).
Scott, P. and Worden, R. (2012). Semantic mapping to
simplify deployment of HL7 v3 Clinical Document
Architecture. Journal of Biomedical Informatics,
45(4):697–702.
Sinaci, A. A. and Erturkmen, G. B. L. (2013). A federated
semantic metadata registry framework for enabling
interoperability across clinical research and care do-
mains. Journal of Biomedical Informatics, 46(5):784
– 794.
Stevens, R. and Sattler, U. (2013). Post-
coordination: Making things up as you go along.
http://ontogenesis.knowledgeblog.org/1305.
Tao, C., Jiang, G., Wei, W., Solbrig, H. R., and Chute, C. G.
(2011). Towards Semantic-Web Based Representation
and Harmonization of Standard Meta-data Models for
Clinical Studies. AMIA Summits on Translational Sci-
ence Proceedings, 2011:59–63.
Ulrich, AK, K., P, D.-H., JK, H., and J, I. (2016). Metadata
repository for improved data sharing and reuse based
on hl7 fhir. Studies in Health Technology and Infor-
matics, 228:162–166.
MODELSWARD 2019 - 7th International Conference on Model-Driven Engineering and Software Development
116
