
A Split-Merge Evolutionary Clustering Algorithm
Veselka Boeva
1
, Milena Angelova
2
and Elena Tsiporkova
3
1
Computer Science and Engineering Dept., Blekinge Institute of Technology, Karlskrona, Sweden
2
Computer Systems and Technologies Dept., Technical University of Sofia, Plovdiv, Bulgaria
3
The Collective Center for the Belgian Technological Industry, Brussels, Belgium
Keywords:
Data Mining, Evolutionary Clustering, Bipartite Clustering, PubMed Data, Unsupervised Learning.
Abstract:
In this article we propose a bipartite correlation clustering technique that can be used to adapt the existing
clustering solution to a clustering of newly collected data elements. The proposed technique is supposed to
provide the flexibility to compute clusters on a new portion of data collected over a defined time period and
to update the existing clustering solution by the computed new one. Such an updating clustering should better
reflect the current characteristics of the data by being able to examine clusters occurring in the considered
time period and eventually capture interesting trends in the area. For example, some clusters will be updated
by merging with ones from newly constructed clustering while others will be transformed by splitting their
elements among several new clusters. The proposed clustering algorithm, entitled Split-Merge Evolutionary
Clustering, is evaluated and compared to another bipartite correlation clustering technique (PivotBiCluster)
on two different case studies: expertise retrieval and patient profiling in healthcare.
1 INTRODUCTION
In this work, we are interested in developing evolu-
tionary clustering techniques that are suited for ap-
plications affected by concept drift phenomena. In
many practical applications such as, expertise (or doc-
ument) retrieval systems the information available in
the system database is periodically updated by col-
lecting (extracting) new data. The available data el-
ements, e.g., experts in a given domain, are usually
partitioned into a number of disjoint subject cate-
gories. It is becoming impractical to re-cluster this
large volume of available information. Profiling of
users with wearable applications with the purpose to
provide personalized recommendations is another ex-
ample. As more users get involved one needs to re-
cluster the initial clusters and also assign new incom-
ing users to the existing clusters. In the context of pro-
filing of machines (industrial assets) for the purpose
of condition monitoring the existing original clusters
can become outdated caused by aging of the machines
and degradation of performance due to influence of
changing external factors. This outdating of models
is in fact a concept drift and requires that the cluster-
ing techniques, used for deriving the original machine
profiles, can deal with such a concept drift and enable
reliable and scalable model update.
Incremental clustering methods process one data
element at a time and maintain a good solution by ei-
ther adding each new element to an existing cluster or
placing it in a new singleton cluster while two existing
clusters are merged into one (Charikar et al., 1997).
Incremental algorithms also bear a resemblance to
one-pass stream clustering algorithms (O’Callaghan
et al., 2001). Although, one-pass stream clustering
methods address the scalability issues of the cluster-
ing problem, they are not sensitive to the evolution of
the data, because they assume that the clusters are to
be computed over the entire data stream.
The clustering scenario discussed herein is differ-
ent from the one treated by incremental clustering
methods. Namely, the evolutionary clustering tech-
niques considered in this work are supposed to pro-
vide the flexibility to compute clusters on a new por-
tion of data collected over a defined time period and
to update the existing clustering solution by the com-
puted new one. Such an updating clustering should
better reflect the current characteristics of the data by
being able to examine clusters occurring in the con-
sidered time period and eventually capture interesting
trends in the area. We propose and study two dif-
ferent clustering algorithms to be suited for the dis-
cussed scenario: PivotBiCluster (Ailon et al., 2011)
and Split-Merge Evolutionary Clustering. Both algo-
Boeva, V., Angelova, M. and Tsiporkova, E.
A Split-Merge Evolutionary Clustering Algorithm.
DOI: 10.5220/0007573103370346
In Proceedings of the 11th International Conference on Agents and Artificial Intelligence (ICAART 2019), pages 337-346
ISBN: 978-989-758-350-6
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
337

rithms are bipartite correlation clustering algorithms
that do not need prior knowledge about the optimal
number of clusters in order to produce a good cluster-
ing solution. In the final clustering generated by the
PivotBiCluster algorithm some clusters are obtained
by merging clusters from both side of the graph, i.e.
some of existing clusters will be updated by some of
the computed new ones. However, existing clusters
cannot be split by the PivotBiCluster algorithm even
the corresponding correlations with clusters from the
newly extracted data elements reveal that these clus-
ters are not homogeneous. This has motivated us to
develop a new Split-Merge Evolutionary Clustering
algorithm that overcomes this disadvantage. Namely,
our algorithm is able to analyze the correlations be-
tween two clustering solutions and based on the dis-
covered patterns it treats the existing clusters in dif-
ferent ways. Thus some clusters will be updated by
merging with ones from newly constructed clustering
while others will be transformed by splitting their el-
ements among several new clusters.
The rest of the paper is organized as follows. Sec-
tion 2 reviews related works. Section 3 briefly dis-
cusses the PivotBiCluster algorithm and describes the
proposed Split-Merge Evolutionary Clustering tech-
nique. Section 4 introduces the two case studies used
to compare and evaluate the two algorithms. Section 5
presents the evaluation of the proposed evolutionary
clustering algorithm in expertise retrieval and patient
profiling in healthcare domains and discusses the ob-
tained results. Section 6 is devoted to conclusions and
future work.
2 RELATED WORK
The model of incremental algorithms for data cluster-
ing is motivated by practical applications where the
demand sequence is unknown in advance and a hier-
archical clustering is required. Incremental clustering
methods process one data element at a time and main-
tain a good solution by either adding each new ele-
ment to an existing cluster or placing it in a new sin-
gleton cluster while two existing clusters are merged
into one (Charikar et al., 1997).
To qualify the type of cluster structure present
in data, Balcan introduced the notion of clusterabil-
ity (Balcan et al., 2008). It requires that every element
be closer to data in its own cluster than to other points.
In addition, Balcan showed that the clusterings that
adhere to this requirement are readily detected offline
by classical batch algorithms. On the other hand, it
was proven by Ackerman (Ackerman and Dasgupta,
2014) that no incremental method can discover these
partitions. Thus, batch algorithms are significantly
stronger than incremental methods in their ability to
detect cluster structure.
Incremental algorithms also bear a resemblance to
one-pass clustering algorithms for data stream prob-
lems (O’Callaghan et al., 2001). Such algorithms
need to maintain a substantial amount of informa-
tion so that important details are not lost. For ex-
ample, the algorithm in (O’Callaghan et al., 2001)
is implemented as a continuous version of k-means
algorithm which continues to maintain a number of
cluster centers which change or merge as necessary
throughout the execution of the algorithm. In ad-
dition, Lughofer proposes a dynamic clustering al-
gorithm which is equipped with dynamic split-and-
merge operations and which is also dedicated to incre-
mental clustering of data streams (Lughofer, 2012).
In (Fa and Nandi, 2012) similarly to the approach of
Lughofer a set of splitting and merging action condi-
tions are defined, where optional splitting and merg-
ing actions are only triggered during the iterative pro-
cess when the conditions are met. Although, one-pass
stream clustering methods address the scalability is-
sues of the clustering problem, they are not sensitive
to the evolution of the data because they assume that
the clusters are to be computed over the entire data
stream.
The clustering scenario discussed herein is differ-
ent from the one treated by incremental clustering
methods. Namely, the evolutionary clustering tech-
nique proposed in this work is supposed to provide
the flexibility to compute clusters on a new portion of
data collected over a defined time period and to up-
date the existing clustering solution by the computed
new one.
Gionis et al. proposed an approach to clustering
that is based on the concept of aggregation (Gionis
et al., 2007). They are interested in a problem in
which a number of different clusterings are given on
some data set of elements. The objective is to produce
a single clustering of the elements that agrees as much
as possible with the given clusterings. Clustering ag-
gregation provides a framework for dealing with a
variety of clustering problems. For instance, it can
handle categorical or heterogeneous data by produc-
ing a clustering on each available attribute and then
aggregating the produced clusterings into a single re-
sult. Another possibility is to combine the results
of several clustering algorithms applied on the same
dataset etc. Clustering aggregation can be thought as
a more general model of multi-view clustering pro-
posed in (Bickel and Scheffer, 2004). The multi-view
approach considers clustering problems in which the
available attributes can be split into two independent
ICAART 2019 - 11th International Conference on Agents and Artificial Intelligence
338

subsets. A clustering is produced on each subset and
then the two clusterings are combined into a single re-
sult. Consensus clustering algorithms deal with sim-
ilar problems to those treated by clustering aggrega-
tion techniques. Namely, such algorithms try to rec-
oncile clustering information about the same data set
coming from different sources (Boeva et al., 2014) or
from different runs of the same algorithm (Goder and
Filkov, 2008). The both clustering techniques are not
suited for our scenario, since they are used to inte-
grate a number of clustering results generated on one
and the same data set.
An interesting split-merge-evolve algorithm for
clustering data into k number of clusters is proposed
by Wang et al. (Wang et al., 2018). The algorithm
randomly divides data into k clusters initially, then re-
peatedly splits bad clusters and merges closest clus-
ters to evolve the final clustering result. This algo-
rithm has the ability to optimize the clustering result
in scenarios where new data samples may be added in
to existing clusters. However, a k cluster output is al-
ways provided by the algorithm, i.e. it is not sensitive
to the evolution of the data, as well.
The idea for the proposed Split-Merge Evolution-
ary Clustering algorithm is inspired by the work of
Xiang et al. (Xiang et al., 2012). They have pro-
posed a split-merge framework that can be tailored
to different applications. The framework models two
clusterings as a bipartite graph which is decomposed
into connected components, and each component is
further decomposed into subcomponents. Pairs of
related subcomponents are then taken into consid-
eration in designing a clustering similarity measure
within the framework.
3 METHODS
3.1 Description of the Framework
Let us formalize the cluster updating problem we
are interested in. We assume that X is the available
set of data points and each data point is represented
by a vector of attributes (features). In addition, the
data points are partitioned into k groups, i.e. C =
{C
1
, C
2
, . . . , C
k
} is an existing clustering solution of X
and each C
i
(i = 1, 2, . . . , k) can be considered as a dis-
joint cluster. In addition, a new set X
0
of recently ex-
tracted data elements (samples) is created, i.e. X ∩ X
0
is an empty set. Each data point in X
0
is again repre-
sented by a list of attributes and C
0
= {C
0
1
, C
0
2
, . . . , C
0
k
0
}
is a clustering solution of X
0
. The objective is to pro-
duce a single clustering of X ∪X
0
by combining C and
C
0
in such a way that the obtained clustering realis-
tically reflects the current distribution in the domain
under interest.
3.2 Pivot Bi-Clustering Algorithm
Two existing clustering techniques are suitable for
the considered context: correlation clustering (Bansal
et al., 2004) and bipartite correlation clustering (Ailon
et al., 2011). The latter algorithm seems to be better
aligned to our clustering scenario. In Bipartite Corre-
lation Clustering (BCC) a bipartite graph is given as
input, and a set of disjoint clusters covering the graph
nodes is output. Clusters may contain nodes from ei-
ther side of the graph, but they may possibly contain
nodes from only one side. A cluster is thought as a
bi-clique connecting all the objects from its left and
right counterparts. Consequently, a final clustering is
a union of bi-cliques covering the input node set. We
compare our Split-Merge correlation clustering algo-
rithm described in the following section with PivotBi-
Cluster realization of the BCC algorithm (Ailon et al.,
2011). The PivotBiCluster algorithm is implemented
according to the original description given in (Ailon
et al., 2011).
Notice that in our considerations the input graph
nodes of the PivotBiCluster algorithm are clusters and
in the final clustering some clusters are obtained by
merging clusters (nodes) from both sides of the graph,
i.e. some of the existing clusters will be updated by
some of the computed new ones. However, existing
clusters cannot be split by the BCC algorithm even
the corresponding correlations with clusters from the
newly extracted data elements reveal that these clus-
ters are not homogeneous.
3.3 Split-Merge Evolutionary
Clustering Algorithm
In this paper, we propose an evolutionary clustering
algorithm that overcomes the above mentioned disad-
vantage of BCC algorithm. Namely, our algorithm is
able to analyze the correlations between two cluster-
ing solutions C and C
0
and based on the discovered
patterns it treats the existing clusters (C) in different
ways. Thus, some clusters will be updated by merg-
ing with ones from newly constructed clustering (C
0
)
while others will be transformed by splitting their ele-
ments among several new clusters. One can find some
similarity between our idea and an interactive cluster-
ing model proposed in (Awasthi et al., 2017). In this
model, the algorithm starts with some initial cluster-
ing of data and the user may request a certain cluster
to be split if it is overclustered (intersects two or more
clusters in the target clustering). The user may also
A Split-Merge Evolutionary Clustering Algorithm
339

request to merge two given clusters if they are under-
clustered (both intersect the same target cluster).
As it was already mentioned in Section 2 our evo-
lutionary clustering algorithm is inspired by a split-
merge framework proposed by Xiang et al. in (Xiang
et al., 2012). By modeling the intrinsic relation be-
tween two clusterings as a bipartite graph, they have
designed a split-merge framework that can be used
to obtain similarity measures to compare clusterings
on different data sets. The problem addressed in this
article is different from the one considered by Xiang
et al. (Xiang et al., 2012). Namely, we concern with
the development of split-merge framework that can be
used to adjust the existing clustering solution to newly
arrived data. Our framework also models two cluster-
ings (the existing and the newly constructed one) as a
bipartite graph which is decomposed into connected
components (bi-cliques) (see Fig. 1 (a), (b) and (c)).
Each component is further analysed and if it is neces-
sary it is decomposed into subcomponents (see Fig. 1
(c) and (d)). The subcomponents are then taken into
consideration in producing the final clustering solu-
tion. For example, if an existing cluster is overclus-
tered (Fig. 1 (b)), i.e. it intersects two or more clus-
ters in the new clustering, it is split between those. If
several existing clusters intersect the same new clus-
ter, i.e. they are underclustered (Fig. 1 (a)), they are
merged with that cluster.
Let us formally describe the proposed Split-Merge
Evolutionary Clustering algorithm. The input bipar-
tite graph is G = (C, C
0
, E), where C and C
0
are sets
of clusters of left and right nodes and E is a subset of
C ×C
0
that represents correlations between the nodes
of two sets. The three main steps of the algorithm are
as follows:
1. Initially, all unreachable nodes from either side of
G are found. These are singleton clusters (out-
liers) in our final clustering solution. We remove
these nodes from the graph.
2. At the second step, all bi-cliques of G are found
and considered. If a bi-clique connects a node
from the left side (C) of G with several nodes from
C
0
the elements of this node have to be split among
the corresponding nodes from C
0
(see Fig. 1 (b)).
In the opposite case, i.e., when we have a bi-clique
that connects a node from the right side (C
0
) of G
with several nodes from left those nodes have to
be merged with that node (cluster) (see Fig. 1 (a)).
All clustered nodes are removed from the graph.
3. At the final step, the remained bi-cliques are de-
composed into split/merge subcomponents. Each
bi-clique, which is a bipartite graph, is trans-
formed into a tripartite graph constructed by two
(split and merge) bipartite graphs. Suppose G
i
=
(C
i
, C
0
i
, E
i
) is the considered bi-clique. Then the
corresponding tripartite graph is built by the fol-
lowing two bipartite graphs: G
iL
= (C
i
, E
i
, E
iL
)
and G
iR
= (E
i
, C
0
i
, E
iR
), where C
i
, C
0
i
and E
i
are
ones from G
i
, E
iL
is a subset of C
i
× E
i
that repre-
sents correlations between the nodes of C
i
and E
i
,
and E
iR
is a subset of E
i
×C
0
i
representing correla-
tions between the nodes of E
i
and C
0
i
(see Fig. 1 (c)
and (d)). For example, c
i
∈ C
i
will be correlated
with all pairs (c
j
, c
0
k
) ∈ E
i
such that c
i
≡ c
j
, and
c
0
i
∈C
0
i
will be correlated with all pairs (c
j
, c
0
k
) ∈ E
i
such that c
0
i
≡ c
0
k
. First all overclustered nodes
of G
iL
are split and new temporary clusters are
formed as a result. Then we perform the corre-
sponding merging for all underclustered nodes in
G
iR
. For example, in Fig. 1 (d), cluster C
2
will
first be split among clusters C
0
1
, C
0
2
and C
0
3
, i.e.
three new clusters, denoted by (C
2
, C
0
1
), (C
2
, C
0
2
)
and (C
2
, C
0
3
), will be obtained. Then at the third
step of the algorithm clusters (C
2
, C
0
1
) and (C
3
, C
0
1
)
will be merged together.
The pseudocode of the proposed Split-Merge Evo-
lutionary Clustering algorithm is given in Algo-
rithm 1. In addition, the algorithm is illustrated with
an example in Fig. 2. The clustering solution gener-
ated by the Split-Merge Clustering is compared to one
produced by the PivotBiCluster. It is interesting to no-
tice that the two algorithms will produce very differ-
ent clustering solutions on the same input graph. For
example, the Split-Merge Clustering will generate a
4-cluster solution while one obtained by the PivotBi-
Cluster will have only 2 clusters. The latter number is
quite low taking into account the number of clusters
in the two input clusterings. Moreover, as it was men-
tioned in the previous section the PivotBiCluster al-
gorithm cannot produce a clustering solution in which
existing clusters are split among new clusters.
4 CASE STUDIES
Luxburg et al. (von Luxburg et al., 2012) argue that
clustering should not be treated as an application-
independent mathematical problem, but should
always be studied in the context of its end-use. Mo-
tivated by this study we have illustrated and initially
evaluated the two studied clustering algorithms in
two different case studies. We have compared the
performance of the algorithms in expertise retrieval
domain by applying them on data extracted from
PubMed repository. In addition, a case study in profil-
ing patients in healthcare domain has been conducted.
ICAART 2019 - 11th International Conference on Agents and Artificial Intelligence
340

Algorithm 1 : Split-Merge Evolutionary Clustering Algo-
rithm.
1: function SPLIT-MERGE(G = (C, C
0
, E))
2: for all nodes c ∈ C ∪C
0
do (*First step*)
3: if c is an unreachable node then
4: Turn c into a singleton and remove it from G
5: end if
6: end for
7: for all nodes c ∈ C ∪C
0
do (*Second step*)
8: Choose c
1
uniformly at random from C
9: if c
1
is the only node from C that takes part in a bi-
clique connecting it with one or several nodes from C
0
then
10: Split c
1
among the corresponding nodes from C
0
11: end if
12: end for
13: for all nodes c ∈ C ∪C
0
do
14: Choose c
0
1
uniformly at random from C
0
15: if c
0
1
is the only node from C
0
that takes part in a bi-
clique connecting it with one or several nodes from C then
16: Merge c
0
1
with the corresponding nodes from C
17: end if
18: end for
19: for all nodes c ∈ C do (*Third step*)
20: Choose c
1
uniformly at random from C
21: Split c
1
among its adjacent nodes from C
0
and form
new temporary clusters
22: end for
23: for all nodes c
0
∈ C
0
do
24: Choose c
0
1
uniformly at random from C
0
25: Merge c
0
1
with its adjacent nodes from the built set of
temporary clusters
26: Remove the clustered nodes from G
27: end for
28: return all connected components (bi-cliques) as clusters
of X ∪X
0
29: end function
4.1 Expertise Retrieval Domain
Currently, organizations search for new employees
not only relying on their internal information sources,
but they also use data available on the Internet to
locate the required experts. Thus the need for ser-
vices that enable finding experts grows especially
with the expansion of virtual organizations. People
are more often working together by forming task-
specific teams across geographic boundaries. The for-
mation and sustainability of such virtual organizations
greatly depends on their ability to quickly trace those
people who have the required expertise. In response
to this, research on identifying experts from on-line
data sources (Abramowicz et al., 2011), (Balog and
de Rijke, 2007), (Bozzon et al., 2013), (Hristoskova
et al., 2013), (Stankovic et al., 2011), (Singh et al.,
2013), (Tsiporkova and Tourw
´
e, 2011),(Boeva et al.,
2016), (Boeva et al., 2018), (Lin et al., 2017) has been
gradually gaining interest in the recent years.
4.1.1 Case Description
Let us suppose that an expertise recommender system
for finding biomedical experts based on on-line data is
under development. The system builds and maintains
Figure 1: Split-Merge Framework: a) a bi-clique that con-
tains underclustered nodes (C
1
and C
2
intersect C
0
1
); b) a
bi-clique that contains an overclustered node (C
1
intersects
C
0
1
, C
0
2
and C
0
3
); c) a bi-clique that has to be decomposed into
subcomponents d) a tripartite graph obtained by decompos-
ing the bi-clique depicted in (c) into split (left) and merge
(right) subcomponents.
a big repository of biomedical experts by extracting
the information about experts’ peer-reviewed articles
that are published and indexed in PubMed. The ex-
perts stored in such big data repositories are usually
partitioned into a number of subject categories in or-
der to facilitate the further search and identification of
experts with the appropriate skills and knowledge. In
addition, the system database is periodically updated
by extracting new data. It is becoming impractical
to re-cluster this large volume of available informa-
tion. Therefore, the objective is to update the existing
expert partitioning by the clustering produced on the
newly extracted experts.
4.1.2 Data Sets
The data needed for our task is extracted from
PubMed, which is one of the largest repositories of
peer-reviewed biomedical articles published world-
wide. Medical Subject Headings (MeSH) is a con-
trolled vocabulary developed by the US National Li-
brary of Medicine for indexing research publications,
articles and books. Using the MeSH terms associated
A Split-Merge Evolutionary Clustering Algorithm
341
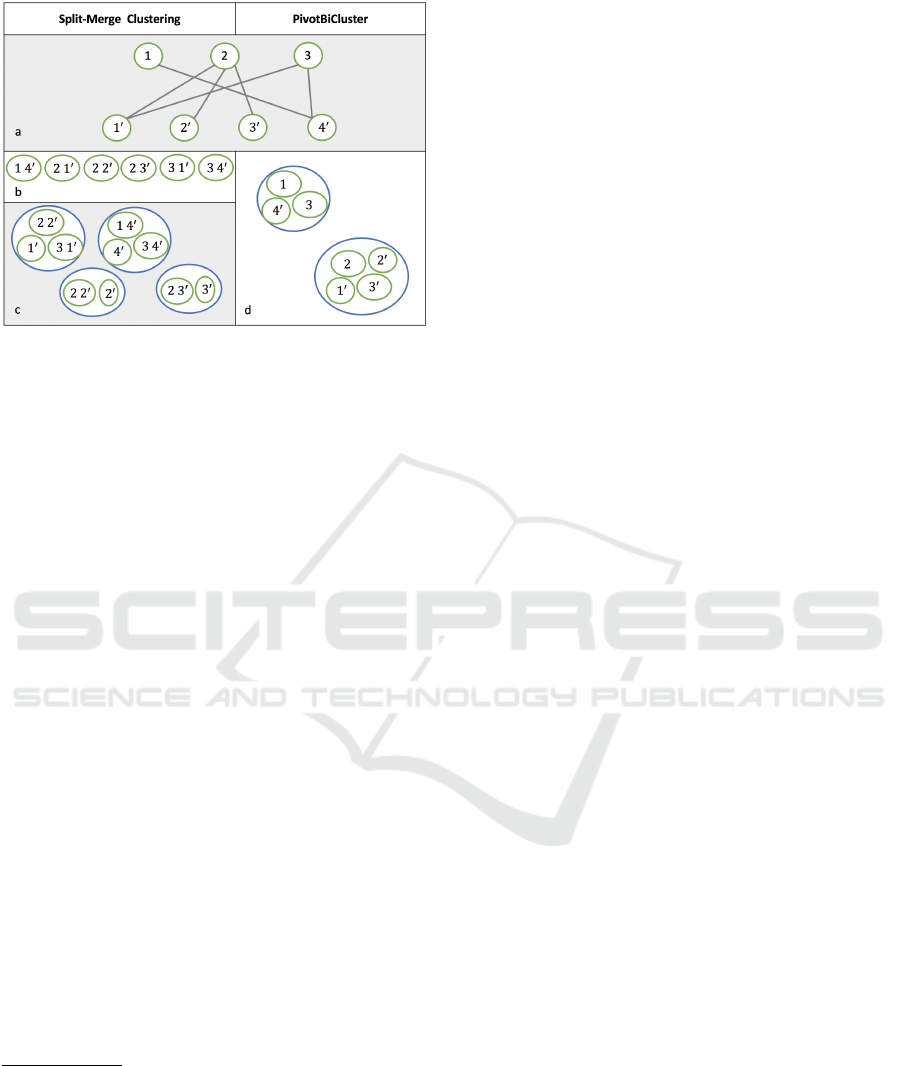
Figure 2: Clustering solutions generated by Split-Merge
Clustering (left) and PivotBiCluster (right), respectively: a)
the input bipartite graph; b) temporary clusters formed by
Split-Merge Clustering after splitting overclustered nodes
from the left (upper) set ({1, 2, 3}) of the graph among cor-
responding nodes from the right (below) set ({1
0
, 2
0
, 3
0
, 4
0
});
c) the final clustering solution produced by Split-Merge
Clustering, d) the final clustering solution produced by Piv-
otBiCluster.
1
with peer-reviewed articles published by the above
considered researchers and indexed in the PubMed,
we extract such authors and construct their expert pro-
files. An expert profile is defined by a list of MeSH
terms used in the PubMed articles of the author in
question to describe her/his expertise areas.
We have extracted a set of 4343 authors from the
PubMed repository. After resolving the problem with
ambiguity
2
the set is reduced to one containing only
3753 different researchers. Then each author is also
represented by a list of all different MeSH headings
used to describe the major topics of her/his PubMed
articles.
In addition to the above set of 3753 biomedical re-
searchers we have used a set of 102 researchers who
have taken part in a scientific conference devoted to
integrative biology
3
. These researchers have been
grouped into 8 clusters with respect to the confer-
ence sessions. They are considered as relevant ex-
perts, thus, used as the ground truth to benchmark the
results of the studied clustering algorithms.
1
A cluster is represented by a circle or an ellipse. An ellipse
with two cluster labels inside, e.g., 2 1
0
, means that some
elements from the first cluster (2) are added to the second
cluster (1
0
).
2
This problem refers to the fact that multiple profiles may
represent one and the same person and therefore must be
merged into a single generalized expert profile.
3
Integrative Biology 2017: 5th International Conference on
Integrative Biology (London, UK, June 19-21, 2017).
4.2 Patient Profiling in Healthcare
Domain
The volumes of current patient data as well as their
complexity make clinical decision making more chal-
lenging than ever for physicians and other care givers.
Decision Support Systems (DSS) can be used to pro-
cess data and form recommendations and/or predic-
tions to assist such decision makers (Belle et al.,
2013). Data mining techniques can be applied to iden-
tify pattern or rules about various quality problems.
For example, profiling together patients who share
similar clinical conditions can facilitate the diagno-
sis and initial treatment of individuals having similar
illness predisposition.
The ability of machine learning and data mining
tools to identify significant features from complex
data sets detects their importance. A variety of such
techniques have already been proposed in healthcare
domain (Cheng et al., 2013), (Aishwarya and Anto,
2014), (Golino et al., 2014), (Menasalvas et al., 2018).
4.2.1 Case Description
Let us suppose a decision support system that can be
used to study and associate the patient anthropometric
measurements with the person increased risk for car-
diovascular disease, e.g., hypertension, is under de-
velopment. The core of the system is based on cluster-
ing techniques which provide groupings of profiles of
individuals with similar anthropometric features, e.g.,
such as body mass index (BMI), waist (WC) and hip
circumference (HC), and waist hip ratio (WHR). The
classification of groups of patients who share proper-
ties in common might provide useful information for
the diagnosis and initial management of risk for hy-
pertension or other cardiovascular disease. For exam-
ple, the patients who share the same profile should
probably have similar predisposition and should be
provided similar healthcare recommendations. In ad-
dition, the system must be able to update and im-
proved the produced anthropometric categories by the
clusters generated on newly arrived patients anthropo-
metric measurements.
4.2.2 Data Sets
The dataset used in this case study is publicly avail-
able and published in (Golino et al., 2014). The data
contains 400 undergraduate students aged between 16
and 63 years old, where a 56.3% are women. The fol-
lowing features describe the data: age, obesity, BMI,
WC, HC, WHR, Systolic Blood Pressure (SBP), Di-
astolic Blood Pressure (DBP), preh for women and
ICAART 2019 - 11th International Conference on Agents and Artificial Intelligence
342

hyper for men, where the preh and hyper are classi-
fication labels that show what kind of blood pressure
the individual has (e.g., regular or hyper). According
to the results published in (Li et al., 2016) people can
be grouped into six clusters depending on their blood
pressure. Based on this the individuals in our test data
set have been grouped into 6 clusters. This grouping
is considered as the ground truth to benchmark the
results generated by the two studied clustering algo-
rithms.
5 EXPERIMENTS AND
DISCUSSION
5.1 Metrics
The data mining literature provides a range of differ-
ent cluster validation measures, which are broadly di-
vided into two major categories: external and inter-
nal (Jain and Dubes, 1988). External validation mea-
sures have the benefit of providing an independent
assessment of clustering quality, since they validate
a clustering result by comparing it to a given exter-
nal standard. However, an external standard is rarely
available. Internal validation techniques, on the other
hand, avoid the need for using such additional knowl-
edge, but have the alternative problem to base their
validation on the same information used to derive the
clusters themselves.
In this work, we have implemented three differ-
ent validation measures for estimating the quality of
clusters, produced by the two studied clustering al-
gorithms. Since we have a benchmark clustering of
the set of 102 biomedical researchers, described in the
foregoing section, we have used the F-measure as an
external validation metric (Larsen and Aone, 1999).
The F-measure is the harmonic mean of the precision
and recall values for each cluster. For a perfect clus-
tering the maximum value of the F-measure is 1. In
addition, Silhouette Index (SI) has been applied as an
internal measure to assess compactness and separa-
tion properties of the generated clustering solutions
(Rousseeuw, 1987). The values of Silhouette Index
vary from -1 to 1.
In addition to the above two metrics, we have used
Jaccard index (Jaccard similarity coefficient) (Jac-
card, 1912) to evaluate the stability of a clustering
method. The Jaccard Index ranges from 0 to 1, where
a higher value indicates a higher similarity between
clustering solutions. Jaccard Index has been used to
measure the similarity between the generated cluster-
ing solutions and the benchmark partitioning of the
data in the second case study.
5.2 Implementation and Availability
We used the Entrez Programming Utilities (E-
utilities) to download all the publications associated
with the extracted biomedical researchers (Sayers,
2010). The E-utilities are the public API to the NBCI
Entrez system and allow access to all Entrez databases
including PubMed, PMC, Gene, Nuccore and Pro-
tein. For calculation of semantic similarities between
MeSH headings, we use MeSHSim which is imple-
mented in an R package. It also supports querying
the hierarchy information of a MeSH heading and
information of a given document including title, ab-
straction and MeSH headings (Zhou and Shui, 2015).
The two studied clustering algorithms (Split-Merge
Clustering and PivotBiCluster) are implemented in
Python. The cluster validation measures (see Sec-
tion 5.1) used to validate the clustering solutions gen-
erated in our experiments are implemented in scikit-
learn library. Scikit-learn is a Python library for data
mining and data analysis. Supplementary information
is available at GitLab
4
.
5.3 Case Study 1
Initially, we use the first built data set that con-
tains 3753 PubMed expert profiles of biomedical re-
searchers. Each expert profile is a vector of subject
keywords describing the expert’s competence. The
researchers of this set are randomly separated in two
sets. The one set contains 2407 experts grouped into
122 clusters by using k-means and the other one has
1346 experts separated into 112 clusters again by ap-
plying k-means. The number of clusters is determined
by clustering each set applying k-means for different
k and evaluating the obtained solutions by SI. The
two clustering algorithms are then executed twice to
integrate the clustering solutions of these two data
sets. The clustering solution produced by the Pivot-
BiCluster has 95 clusters while the proposed Split-
Merge Clustering algorithm has generated a solution
with 104 clusters. The generated clustering solutions
are evaluated by SI and the average scores are -0.158
(PivotBiCluster) and 0.058 (Split-Merge Clustering),
respectively. Evidently, the Split-Merge Clustering
algorithm outperforms PivotBiCluster on this data set.
We believe this is due to the fact that it adjusts better
to data by being able not only to merge those clusters
4
https://gitlab.com/machine learning vm/clustering
techniques
A Split-Merge Evolutionary Clustering Algorithm
343
that are underclustered but also to split those that are
overclustered.
Next, the benchmark set of 102 different expert
profiles is used to generate 10 test data sets couples.
Each test couple separates the researchers randomly
in two sets. The one set (containing 70 experts) of
each couple presents the available set of experts, and
another one (32 experts) is the set of newly extracted
experts. In that way, 10 test clustering couples are
created.
We have studied two different experiment scenar-
ios. In the first scenario the experts in each test set are
grouped into clusters of experts with similar expertise
based on the conference session information, i.e. each
set is partitioned into 8 clusters. In the second sce-
nario for each data sets the optimal number of clusters
is determined by clustering the set applying k-means
for different k and evaluating the obtained solutions
by SI. In this way, two different experiments have
been conducted on 10 test data set couples. In both
experiments, the PivotBiCluster algorithm is executed
10 times (i.e., 100 executions in total for each ex-
periment) to integrate the corresponding clusterings.
In comparison to the PivotBiCluster, the Split-Merge
Clustering is conducted only once on each test cou-
ple, since it does not start by a random cluster selec-
tion. Namely, it initially identifies those bi-cliques
that have to be split and merged, respectively, i.e. the
clustering result is not dependent on the algorithm ini-
tialization.
The obtained results for SI and F-measure are
shown in Table 1 and Table 2, respectively. Observe
that the PivotBiCluster outperforms the Split-Merge
Clustering algorithm only in one case. Namely, it has
generated a higher F-measure value than the Split-
Merge Clustering algorithm in the first experiment.
It is interesting to notice that in the second experi-
ment (see Table 2) the SI scores are not only higher
in comparison to the ones generated in the first exper-
iment, but they are also positive. Evidently, using the
optimal number of clusters significantly improves the
quality of the generated clustering solutions with re-
spect to compactness and separation properties. How-
ever, the corresponding F-measure scores are lower
than the ones generated in the first experiment.
The above results support the mentioned above ar-
guments of Luxburg et al. (von Luxburg et al., 2012)
that the cluster evaluation methods can produce con-
tradictory results and often do not serve their purpose.
The main point of the authors is that clustering al-
gorithms cannot be evaluated in a problem indepen-
dent way, i.e. the known cluster validation measures
cannot be used to evaluate the usefulness of the clus-
tering. It is still not clear how we can measure the
usefulness of a newly developed clustering algorithm.
Certainly, the proposed Split-Merge Clustering algo-
rithm needs further evaluation and validation in case
studies from different application domains. Thus in
the next section we present an additional evaluation
of the two studied algorithms in a case study from
healthcare domain.
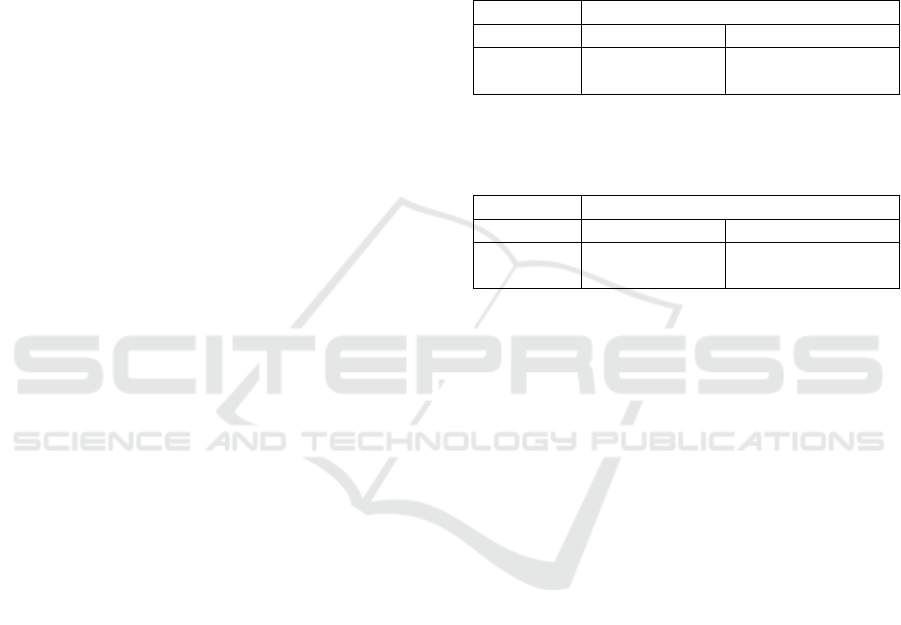
Table 1: Experiment 1: Average F-measure and SI values
generated on the clustering solutions of the 10 test data set
couples.
Experiment 1
Metrics PivotBiCluster Split-Merge Clust.
F-measure 0.618 0.582
SI -0.145 -0.129
Table 2: Experiment 2: Average F-measure and SI values
generated on the clustering solutions of the 10 test data set
couples.
Experiment 2
Metrics PivotBiCluster Split-Merge Clust.
F-measure 0.321 0.331
SI 0.137 0.157
5.4 Case Study 2
In this case study, we have used the data set explained
in Section 4.2.2. This set consists of 400 individ-
ual profiles and it is used to generate 10 test data
set couples by randomly separating the individuals
in two sets. The one set (280 patients) of each cou-
ple presents the available set of individual profiles,
and another one (120 individuals) is the set of newly
collected patients’ profiles. In that way similar to
the first case study we have created 10 test data set
couples. Notice that each patient profile is a vector
of the patient’s anthropometric features (BMI, WC,
HC, WHR), and the patient’s Systolic Blood Pres-
sure (SBP) and Diastolic Blood Pressure (DBP). The
patients’ profiles of each set have been grouped in
6 clusters according to (Li et al., 2016), see Sec-
tion 4.2.2. Namely, the individuals have been grouped
in six clusters depending on their blood pressure. The
obtained clusters are presented by their centroids.
Analogously to the first case study, the PivotBi-
Cluster has been executed ten times for each test data
set couple (i.e., 100 executions in total). The algo-
rithm considers clusters in random order and gener-
ates a different clustering solution for each execu-
tion. As a result, the average value over these ten
executions has been calculated. The above random-
ness is not presented in the Split-Merge Clustering al-
gorithm. Therefore, it has been executed only once
ICAART 2019 - 11th International Conference on Agents and Artificial Intelligence
344

over each test data set couple. Both algorithms are
explained in more detail in Section 3.
The generated clustering solutions are again eval-
uated by SI and F-measure. The obtained average SI
scores are -0.013 (PivotBiCluster) and -0.170 (Split-
Merge Clustering), respectively. Evidently, the Pivot-
BiCluster outperforms the Split-Merge Clustering al-
gorithm with respect to this evaluation criteria. The
results obtained by F-measure also support the better
performance of PivotBiCluster (0.71 against 0.46 for
Split-Merge Clustering) on this data set. However, it
is interesting to observe that the number of clusters of
the clustering solutions generated by the PivotBiClus-
ter on the test data set couples varies from 1 to 5 while
in the case of the Split-Merge Clustering the individ-
uals are grouped into 5 or 6 clusters. Notice that the
benchmark clustering (Section 4.2.2) has 6 clusters.
The above results have motivated us to use Jac-
card Index for an additional comparison of the two
studied algorithms. Namely, we have applied the Jac-
card Index to measure the similarity between the gen-
erated clustering solutions and the benchmark clus-
tering. The corresponding values are 0.081 (Pivot-
BiCluster) and 0.291 (Split-Merge Clustering), i.e.,
the Split-Merge Clustering algorithm has generated a
higher average Jaccard score than the PivotBiCluster.
In addition, we have evaluated the two clustering
algorithms with respect to the purity of the gener-
ated clustering solutions. For this purpose we con-
sider how the two main classes (regular and hyper
blood pressure) are distributed among the clusters.
The score obtained for the benchmark clustering is
0.16. The values generated for the two studied al-
gorithms are 0.025 (PivotBiCluster) and 0.17 (Split-
Merge Clustering), respectively. Evidently, the Split-
Merge Clustering performs better than PivotBiCluster
with respect to this criteria and manages to preserve
a level of purity closed to one of the benchmark clus-
tering.
6 CONCLUSION AND FUTURE
WORK
In this work, we have proposed a novel evolutionary
clustering technique, entitled Split-Merge Evolution-
ary Clustering, that can be used to adapt the existing
clustering solution to a clustering of newly collected
data elements. The proposed technique has been com-
pared to PivotBiCluster, an existing clustering algo-
rithm that is also suitable for concept drift scenarios.
The two algorithms have been evaluated and demon-
strated in two different case studies. The Split-Merge
Clustering algorithm has shown better performance
than the PivotBiCluster in most of the studied experi-
mental scenarios.
For future work, we aim to pursue further compar-
ison and evaluation of the two clustering algorithms in
different application domains and on richer data sets.
REFERENCES
Abramowicz, W., Bukowska, E., Dzikowski, J., Filipowska,
A., and Kaczmarek, M. (2011). Semantically en-
abled experts finding system - ontologies, reasoning
approach and web interface design. In ADBIS 2011,
Research Communications, Proc. II of the 15th East-
European Conference on Advances in Databases and
Information Systems, September 20-23, Vienna, Aus-
tria, pages 157–166.
Ackerman, M. and Dasgupta, S. (2014). Incremental clus-
tering: The case for extra clusters. In Proceedings of
the 27th International Conference on Neural Informa-
tion Processing Systems - Volume 1, NIPS’14, pages
307–315.
Ailon, N., Avigdor-Elgrabli, N., Liberty, E., and van
Zuylen, A. (2011). Improved approximation algo-
rithms for bipartite correlation clustering. In Algo-
rithms - ESA 2011 - 19th Annual European Sympo-
sium, Saarbr
¨
ucken, Germany, September 5-9, 2011.
Proceedings, pages 25–36.
Aishwarya, A. and Anto, S. (2014). A medical decision
support system based on genetic algorithm and least
square support vector machine for diabetes disease di-
agnosis. International Journal of Engineering Sci-
ences & Research Technology, 3(4):4042–4046.
Awasthi, P., Balcan, M. F., and Voevodski, K. (2017). Lo-
cal algorithms for interactive clustering. Journal of
Machine Learning Research, 18(3):1–35.
Balcan, M.-F., Blum, A., and Vempala, S. (2008). A
discriminative framework for clustering via similar-
ity functions. In Proceedings of the Fortieth Annual
ACM Symposium on Theory of Computing, STOC ’08,
pages 671–680.
Balog, K. and de Rijke, M. (2007). Finding similar experts.
In Proceedings of the 30th Annual International ACM
SIGIR Conference on Research and Development in
Information Retrieval, SIGIR ’07, pages 821–822.
Bansal, N., Blum, A., and Chawla, S. (2004). Correlation
clustering. Machine Learning, 56(1-3):89–113.
Belle, A., Kon, M. A., and Najarian, K. (2013). Biomedical
informatics for computer-aided decision support sys-
tems: A survey. The Scientific World Journal, pages
1–8.
Bickel, S. and Scheffer, T. (2004). Multi-view clustering.
In Proceedings of the Fourth IEEE International Con-
ference on Data Mining, ICDM ’04, pages 19–26.
Boeva, V., Angelova, M., Boneva, L., and Tsiporkova, E.
(2016). Identifying a group of subject experts using
formal concept analysis. In 8th IEEE International
Conference on Intelligent Systems, IS 2016, Sofia,
Bulgaria, September 4-6, IS IEEE, pages 464–469.
A Split-Merge Evolutionary Clustering Algorithm
345

Boeva, V., Angelova, M., Lavesson, N., Rosander, O., and
Tsiporkova, E. (2018). Evolutionary clustering tech-
niques for expertise mining scenarios. In Proceedings
of the 10th International Conference on Agents and
Artificial Intelligence, ICAART, Volume 2, Funchal,
Madeira, Portugal, January 16-18, pages 523–530.
Boeva, V., Tsiporkova, E., and Kostadinova, E. (2014).
Analysis of Multiple DNA Microarray Datasets, pages
223–234. Springer Berlin Heidelberg.
Bozzon, A., Brambilla, M., Ceri, S., Silvestri, M., and
Vesci, G. (2013). Choosing the right crowd: Ex-
pert finding in social networks. In Proceedings of the
16th International Conference on Extending Database
Technology, EDBT ’13, pages 637–648. ACM.
Charikar, M., Chekuri, C., Feder, T., and Motwani, R.
(1997). Incremental clustering and dynamic informa-
tion retrieval. In Proc. of the 29th Annual ACM Sym-
posium on Theory of Computing, STOC ’97, pages
626–635.
Cheng, C. W., Chanani, N., Venugopalan, J., Maher, K., and
Wang, M. D. (2013). An icu clinical decision support
system using association rule mining. Translational
Engineering in Health and Medicine, IEEE, 1(2):8–
17.
Fa, R. and Nandi, A. K. (2012). Smart: Novel self splitting-
merging clustering algorithm. In European Signal
Processing Conference, Bucharest, Romania, August,
27-32. IEEE.
Gionis, A., Mannila, H., and Tsaparas, P. (2007). Clustering
aggregation. ACM Transaction of Knowledge Discov-
ery Data, 1(1).
Goder, A. and Filkov, V. (2008). Consensus clustering al-
gorithms: Comparison and refinement. In ALENEX,
pages 109–234.
Golino, H. F., de Brito Amaral, L. S., Duarte, S. F. P., and
et al. (2014). Predicting increased blood pressure us-
ing machine learning. Journal of Obesity, 2014.
Hristoskova, A., Tsiporkova, E., Tourw
´
e, T., Buelens, S.,
Putman, M., and Turck, F. D. (2013). A graph-based
disambiguation approach for construction of an expert
repository from public online sources. In ICAART
2013 - Proceedings of the 5th International Confer-
ence on Agents and Artificial Intelligence, Volume 2,
Barcelona, Spain, 15-18 February, pages 24–33.
Jaccard, P. (1912). The distribution of flora in the alpine
zone. New Phytologist, 11:37–50.
Jain, K. A. and Dubes, C. R. (1988). Algorithms for Clus-
tering Data. Prentice-Hall, Inc.
Larsen, B. and Aone, C. (1999). Fast and effective text min-
ing using linear-time document clustering. In Proc.
of the 5th ACM SIGKDD International Conference
on Knowledge Discovery and Data Mining, KDD’99,
pages 16–22. ACM.
Li, Y., Feng, X., Zhang, M., Zhou, M., Wang, N., and
Wangb, L. (2016). Clustering of cardiovascular be-
havioral risk factors and blood pressure among people
diagnosed with hypertension: a nationally representa-
tive survey in china. Sci Rep., 6:27627.
Lin, S., Hong, W., Wang, D., and Li, T. (2017). A survey
on expert finding techniques. Journal of Intelligent
Information Systems, 49:255–279.
Lughofer, E. (2012). A dynamic split-and-merge approach
for evolving cluster models. Evolving Systems, 3:135–
151.
Menasalvas, E. R., Tu
˜
nas, M. J., Bermejo, G., Gonzalo,
C. M., Rodr
´
ıguez-Gonz
´
alez, A., Zanin, M., Pedro, C.
G. D., M
´
endez, M., Zaretskaia, O., Rey, J., Parejo,
C., Bermudez, L. J. C., and Provencio, M. (2018).
Profiling lung cancer patients using electronic health
records. Journal of Medical Systems, 42:1–10.
O’Callaghan, L., Mishra, N., Meyerson, A., Guha, S., and
Motwani, R. (2001). Streaming-data algorithms for
high-quality clustering. In Proceedings of IEEE In-
ternational Conference on Data Engineering, pages
685–694.
Rousseeuw, P. J. (1987). Silhouettes: A graphical aid to
the interpretation and validation of cluster analysis.
Journal of Computational and Applied Mathematics,
20:53–65.
Sayers, E. (2010). A General Introduction to the E-utilities.
In: Entrez Programming Utilities Help [Internet].
Bethesda (MD): National Center for Biotechnology
Information (US).
Singh, H. S., Singh, R., Malhotra, A., and Kaur, M. (2013).
Developing a biomedical expert finding system using
medical subject headings. In Healthcare informatics
research, 19(4): 243–249.
Stankovic, M., Jovanovic, J., and Laublet, P. (2011). Linked
data metrics for flexible expert search on the open
web. In Proceedings of the 8th Extended Semantic
Web Conference on The Semantic Web: Research and
Applications - Volume Part I, ESWC’11, pages 108–
123. Springer-Verlag.
Tsiporkova, E. and Tourw
´
e, T. (2011). Tool support
for technology scouting using online sources. In
Advances in Conceptual Modeling. Recent Develop-
ments and New Directions, pages 371–376. Springer
Berlin Heidelberg.
von Luxburg, U., Williamson, R. C., and Guyon, I. (2012).
Clustering: Science or art? In Proceedings of ICML
Workshop on Unsupervised and Transfer Learning,
volume 27 of Proceedings of Machine Learning Re-
search, pages 65–79.
Wang, M., Huang, V., and Bosneag, A.-M. C. (2018). A
novel split-merge-evolve k clustering algorithm. In
IEEE 4th International Conference on Big Data Com-
puting Service and Applications (BigDataService),
Bamberg, Germany, March 26-29.
Xiang, Q., Mao, Q., Chai, K. M. A., Chieu, H. L., Tsang,
I. W., and Zhao, Z. (2012). A split-merge framework
for comparing clusterings. In ICML, pages 1055-
1062.
Zhou, J. and Shui, Y. (2015). MeSHSim: MeSH(Medical
Subject Headings) Semantic Similarity Measures. R
package version 1.4.0.
ICAART 2019 - 11th International Conference on Agents and Artificial Intelligence
346
