
Quality Management for Big 3D Data Analytics: A Case Study of
Protein Data Bank
Hind Bangui
1,2
, Mouzhi Ge
1,2
and Barbora Buhnova
1,2
1
Institute of Computer Science, Masaryk University, Brno, Czech Republic
2
Faculty of Informatics, Masaryk University, Brno, Czech Republic
Keywords: Big Data, 3D Data Quality, Data Cleaning, 3D Data Processing, Data Analytics, Protein Data Bank.
Abstract: 3D data have been widely used to represent complex data objects in different domains such as virtual reality,
3D printing or biological data analytics. Due to complexity of 3D data, it is usually featured as big 3D data.
One of the typical big 3D data is the protein data, which can be used to visualize the protein structure in a 3D
style. However, the 3D data also bring various data quality problems, which may cause the delay, inaccurate
analysis results, even fatal errors for the critical decision making. Therefore, this paper proposes a novel big
3D data process model with specific consideration of 3D data quality. In order to validate this model, we
conduct a case study for cleaning and analyzing the protein data. Our case study includes a comprehensive
taxonomy of data quality problems for the 3D protein data and demonstrates the utility of our proposed model.
Furthermore, this work can guide the researchers and domain experts such as biologists to manage the quality
of their 3D protein data.
1 INTRODUCTION
Nowadays, the research on three-dimension (3D) data
has been attracting increasing attentions from
research communities and industry. Since the success
of virtual reality development, 3D application has
proven its effectiveness for not only presenting
essential information in vast amounts of data but also
driving complex analyses to explore how these data
interact in a sophisticated environment. There are
various applications of 3D data that reflect the
advantages of 3D techniques such as (Bio) analytical
chemistry enabled by 3D printing (Palenzuela et al.,
2018, real-time vision-based 3D camera tracking
(Crivellaro et al., 2018), and 3D protein structure
prediction problem (Correa et al., 2018).
Nevertheless, there are several negative 3D data
aspects that prevent the research and industry from
reaching reliable results based on the application of
3D techniques, such as getting low-quality 3D results.
Several approaches have been proposed to address
these anomalies and ensure the scalability of 3D
results, such as the reconstruction of 3D data in real-
time for correcting data errors (Dai et al., 2017).
However, these approaches usually require iterations
for re-processing and correcting the 3D model errors.
However, the iteration time is a critical parameter in
some domains like heathcare that requires a timely
precision, quick decision and visualization of patient
information for determining the adequate cures. That
means low-quality data might influence the quality of
drugs and medical equipment. Similarly, the
completeness, correctness and good presentation of
biological science data are required by scientists to
understand the human health background, such as the
formation of the tumours. Accordingly, the cleaning
process of data is an important step for ensuring high-
quality 3D data analysis results.
The Big Data analytics presents new research
opportunities to tackle the 3D data challenges,
particularly, as a large number of open datasets is
freely available for use without any restriction, the
open 3D data play an important role in creating new
innovative public services in different areas.
Accordingly, open 3D data have become a potential
resource for many domains, such as bioinformatics
and virtual reality. Furthermore, the bioinformatics
field focuses on combining the biological data and
computational intelligence approaches, including
machine learning techniques, for processing,
analyzing, and extracting the most considerable
biological information.
286
Bangui, H., Ge, M. and Buhnova, B.
Quality Management for Big 3D Data Analytics: A Case Study of Protein Data Bank.
DOI: 10.5220/0007717402860293
In Proceedings of the 4th International Conference on Internet of Things, Big Data and Security (IoTBDS 2019), pages 286-293
ISBN: 978-989-758-369-8
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

The analysis of 3D data is very complicated for
the traditional data processing applications to deal
with the data (Corral-Corral et al., 2015).
Consequently, there are many challenges when
addressing 3D data problem in term of Big Data
technologies, such as protein data storage, protein
data search, protein data cleaning, protein data
sharing, protein data visualization, and protein data
analysis, whereby 3D data quality is a critical factor
in the data processing because 3D data quality can
directly affect the quality of the analytics results.
This paper therefore proposes a Big Data
processing model by considering the data quality
assessment and improvement. To validate this model,
we further conduct a case study for the protein 3D
data to show the importance of data quality
management in 3D data analytics. When merging the
open 3D data and Big Data technologies, we intend to
address the data quality issues in the whole data
process. This paper is not focused on the data
analytics process rather how to control the data
quality during the data analytics.
The remainder of the paper is structured as
follows. Section 2 provides a brief overview of 3D
data features and application of Big Data technolgies
in 3D data. Section 3 proposes a general big 3D data
processing model. In order to validate this mode,
Section 4 conducts an experiment to process the 3D
protein data. Finally, Section 5 concludes the paper
and outlines the future research.
2 RELATED WORK
Big Data analytics tools have been widely used in
addressing 3D data challenges, espcically in the
biological 3D data analytics. For example, machine
learning approaches have been used in protein 3D
structure prediction. Furthermore, we found that
many research works focus on using the Big Data
analytics in bioinformatics domain. For example, in
(Corral-Corral et al., 2015), the authors have
represented each protein structure by its local
residues. Then, they have developed a method for
constructing a compact vector space model of protein
fold space. Furthermore, they have focused on
developing a model that is learnable by any machine-
learning approach, notably, the centroid clustering
algorithms (i.e., K-means clustering) that are used as
the most confident protein prediction because they
can be even closer to the native structure.
Since 3D data appear to be prevalent in
bioinformatics research, 3D data analytics is one of
the research foci in biological research (Aydin et al.,
2018). Therefore, we consider that the 3D data
analytics in bioinformatics is a typical scenario for
understanding the 3D data analytics. For example,
protein structure prediction is one of the main issues
of the bioinformatics research area that allow
predicting and studying a variety of protein structural
features, including secondary structure, natively
disordered regions, and protein domain boundaries.
Generally, the prediction process can be decomposed
on four different dimensions, which are: Structural
features along the primary sequence of amino acids
(1D), spatial relationships between amino acids (2D),
the tertiary structure of a protein (3D), and the
quaternary structure of a multi-protein complex (4D).
There are different works that use Big Data
technologies to address the biological data
challenges. In (Kalaivani et al., 2018), k-means
algorithm has been used as an efficient algorithm for
protein complex detection. Another work in (Tripathy
et al., 2018) has combined c-means with fuzzy and
spectral clustering for developing a computational
approach for mining cholesterol and their potential
target against GPCR seven helices. On the other hand,
there are some works that have focused on developing
tools that integrate various clustering algorithms such
as iFeature (Chen et al., 2018) that is a python
package and web server for features extraction and
selection from protein and peptide sequences. It
integrates features selection analysis based on k-
means, hierarchical, mean shift, affinity propagation,
DBSCAN clustering algorithms to facilitate the
interpretation of the results for non-expert users.
However, the existence of various clustering
algorithms, which provides different o utput results,
is another problem for researchers in determining
which of hem is the most suitable for further
processing 3D visualization proteins (Vignesh et al.,
2018).
In addition to clustering, supervised learning
techniques (i.e., k-nearest neighbor algorithm) have
been used to assess the quality of a protein model
directly. In (Li et al., 2018), an improved k-nearest
neighbor (KNN) method was used to diagnose breast
cancer. Similarly, in (Tiwari et al., 2018), fuzzy-k-
nearest neighbor was used as a classifier for solving
the issue of efficient classification of nuclear receptor
(a class of protein) and their subfamilies, which plays
an important role in the detection of various human
diseases such as inflammatory diseases and their
related drug design and discovery. Equally, the KNN
smoothing algorithm has been used for classifying
single-cell RNA-Seq data and reducing noise by
aggregating information from similar cells
(neighbors) in a computationally efficient and statisti-
Quality Management for Big 3D Data Analytics: A Case Study of Protein Data Bank
287
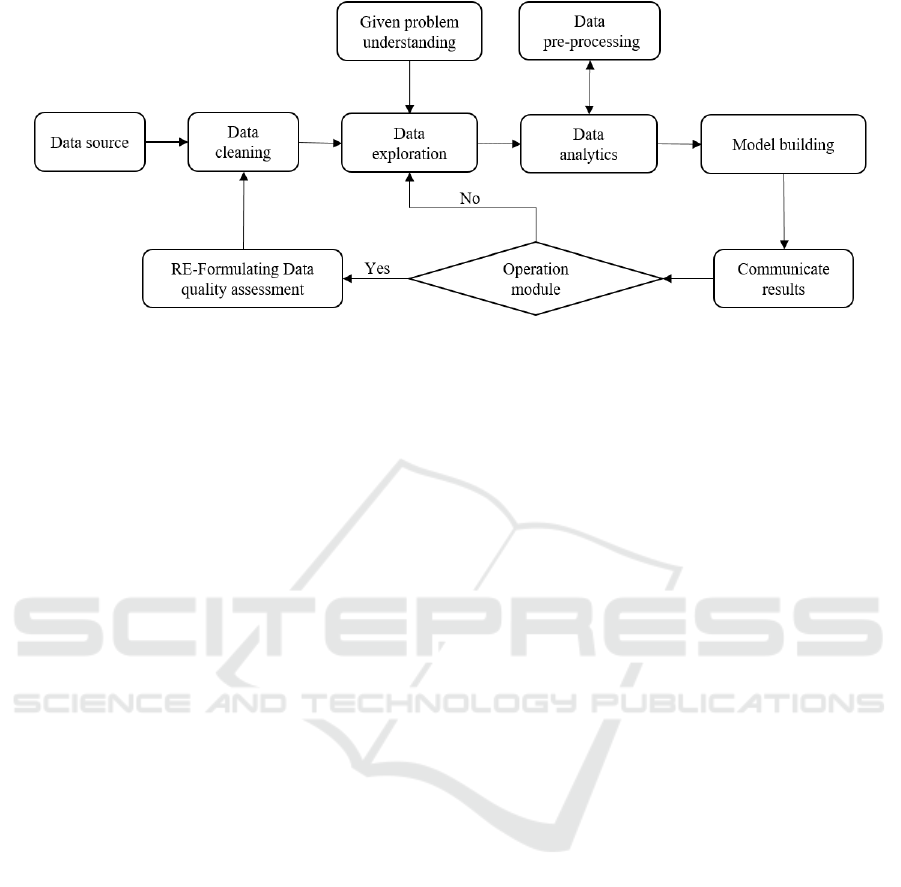
Figure 1: Big 3D Data processing model with data quality management.
cally tractable manner. Accordingly, the supervised
and unsupervised machine learning methods have
been applied to tackle the protein structure problems
and have significantly contributed to advancing
various biological topics. However, the most
supervised and unsupervised works don’t discuss the
cleaning process of protein data before addressing a
given 3D protein problem. Further, they just focus on
proving how to apply directly the Big Data analytics
techniques and use their benefits for facing the 3D
protein challenges. Thus, in the next section, we are
going to conduct an overview regarding the Protein
Data Bank dataset to supervise the validation of 3D
protein data.
In Big Data analytics, data quality is one of the
foundamental aspects because the quality of analytics
result depends on the quality of input data (Ge and
Helfert, 2008). In order to measure data quality,
Wang and Strong (1996) used an exploratory factor
analysis to derive 15 data quality dimensions, which
are widely accepted in the following data quality
research. To organize these data quality dimensions,
Wang and Strong (1996) classified data quality into 4
categories: intrinsic, contextual, representational and
accessibility. Intrinsic information quality consists of
context-independent dimensions such as
believability, accuracy, objectivity and reputation. In
contrast to intrinsic data quality, contextual data
quality highlights the dimensions, which need to be
considered in an application context. Value-added,
relevance, timeliness, completeness and appropriate
amount of data are in this category. Representational
data quality concerns whether the data is presented in
an easily interpretable, understandable, concise and
consistent way. Therefore, the related dimensions are
interpretability, ease of understanding, consistency
and concise representation. Accessibility data quality
emphasizes that the data needs to be accessible yet
still secure, and thus accessibility and security are in
this category (Helfert et al. 2009). In this paper, this
work serves as a foundational work for our
experiment to classify the 3D data quality issues into
quality dimensions.
3 BIG 3D DATA PROCESSING
MODEL
In this section, we present a new model for processing
the big 3D data with specific consideration of data
quality issues in Figure 1. The detailed specification
of each step is explained as follows.
▪ Data Source: It represents the original data stored
that will be used by the data exploration module
for treating a specific problem.
▪ Data Cleaning: It consists of different methods
used to detect the data anomalies (i.e., errors) and
evaluate the quality of data.
▪ Data Exploration: This module aims to extract a
meaningful part or whole data according to the
requirements of a given problem.
▪ Data Pre-processing: It consists of the re-
evaluation of the extracted data quality to
determine useful insights for better decision-
making.
▪ Data Analytics: It consists of different methods
used for analyzing and processing data like
clustering algorithms.
▪ Model Building: It represents the value of the
processed data.
▪ Communicate Results: It generates different
reports concerning the results and errors that
could be used for optimizing the extraction of data
and improving the data quality.
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
288

▪ Operationalizing Module: It filters the results
generated by the communicate results module.
▪ Re-formulating Data Quality Assessment: It
updates the data cleaning module to optimize
further the offered functionalities for ensuring the
data quality.
After storing the data source, the data cleaning
module refers to data quality assessment that is used
to clean and produce high-quality data (Ge and
Helfert 2006). It is first applied to adjust and correct
the data source such as removing the redundancy or
irrelevant data. Then, the data exploration phase
extracts meaningful information according to a given
problem. After that, the data analytics module starts
to process the collected data, at the same time, the pre-
processing data module supervises the process of data
to make sure whether the extracted data need to be
cleaned or not. Next, the model building is achieved
following by the communicate results module. This
latter describes well the quality of the finding results
that will be filtered by the operationalizing module
for finding new criteria not defined in the data
cleaning module. This step that consists of updating
the data assessment is necessary to ensure the high
quality of future results. Furthermore, it helps the Big
Data services (i.e., Big Data analytics) to meet the
modern demands of relevant information processing
and hence decision-making in different domains.
Accordingly, the proposed model can guarantee that
the data cleaning respect to the requirement of a
specific problem. Also, it can ensure that the data
processing will be achieved with the minimum errors.
Therefore, the goal of re-adjusting the data cleaning
criteria is to detect, eliminate and correct errors
arising from the input data, which is the primary
cause of wrong results. Thus, the proposed model
could improve data quality and perform the basic Big
Data lifecycle.
4 CASE STUDY OF PROTEIN
DATA BANK
In this section, we conduct a case study on the PDB
database
1
that is established in 1971. It is a global
representative archive of experimentally determined
3D structures of biological macromolecules. In 2018,
it contains so far 135,536 protein structures. The
proteins databases are classified into two categories:
The primary databases that contain enormous
amounts of protein data submitted directly by the
researchers (i.e., Protein Data Bank for 3D structures
of biological macromolecules), whereas the
secondary databases include the analysis of primary
databases and their updating rates are relatively slow
(i.e. CATH for Protein structure classification).
The Protein Data Bank (http://www.rcsb.org/pdb)
contains one of the biggest open biological archives
used by bioinformatics researchers to study the
biological topics like the 3D protein folding similarity
problem (Berman et al., 2014). This latter aims at
recognizing the proteins that have structural
similarities to other proteins. For that reason, the
biological databases are used to extract the protein
information to evaluate and detect the 3D structural
similarity of given proteins, which could help, for
example, to discover new cures in the healthcare
domain. However, the primary objectives of
biological databases are not only to store, organize
and share data in a structured and searchable manner
with the aim to facilitate 3D data retrieval and
visualization for humans, but also to encourage the
researchers to contribute in these open databases for
supporting further the life sciences. As a result, the
protein databases will be an important source of data
for developing new bio-applications. Accordingly,
the study of the gigantic volumes of open biological
data is necessary in Big Data paradigm for getting
more reliable results in the biological domain and its
related domains.
4.1 Overview of Big Data Applications
in PDB
We firstly pay particular attention to conduct an
overview of the application of Big Data tools for
managing and processing PDB data. We have
collected 14 papers published between 2004 and 2018
that focus on formally describing the selected PDB
database and its related challenges and issues.
Furthermore, we used specific keywords
characterizing Big Data to see how its tools or
concepts are applied to ensure the quality of PDB
data, such as search, indexation, annotation, cleaning,
classification, prediction, representation, protein,
PDB, protein data bank, quality, validation,
compression, analytics, filtering, gathering,
generation, replication. Thus, we have classified the
results in Table 1 to describe the quality issues in the
PDB database.
4.2 Data Quality Issues in PDB
After deriving an overview of the applications of Big
Data for managing the PDB files as Table 1, we have
classified the data quality issues in Table 2 according
to the data quality dimensions, whereby the accuracy
Quality Management for Big 3D Data Analytics: A Case Study of Protein Data Bank
289

dimension refers to how closely the measured PDB
data values are to the real-life event, the completeness
that measures the missing PDB values, and the
consistency that represents the respect of PDB data
constraints. This classification is to facilitate the data
quality management in the process of PDB data
analytics.
PDB database is constructed from structural text
represented as a PDB file. The biologists usually
describe the new PDB information and then they
upload the new text file in the online PDB database.
There is a phase for validating the PDB file before the
online publication. For example, Read et al., (2011)
have proposed to use a report to include a summary
of global quality indicators that can allow others to
judge whether specific conclusions are justified by
the quality of the newly uploaded PDB data.
Similarly, in (Joosten et al., 2014), the PDB_REDO
server is used to refine and validate data submitted to
the online PDB platform. However, after using the
stored PDB data, the output results might be incorrect
because the original data have incorrect and missing
labels.
Several solutions have suggested to tackle the
anomaly by developing new algorithms to allow
automatic rebuilding and remodeling of
crystallographic structures in the Protein Data Bank
(Joosten et al., 2011). Also, in (Van Beusekom et al.,
2018), the PDB-REDO pipeline is exploited for
reannotating and re-refining the glycoprotein
structure models from the PDB database. Similarly,
the PDBFINDER database has been developed as a
searchable PDB entry meta-data to manage quality
information about the PDB entries. Furthermore, all
the PDB errors are stored in PDBREPORT database
for detecting these PDB data anomalies. Another
database called WHY_NOT has been proposed to
explain why PDB entries in any databank do not exist
(Touw et al., 2014). However, these suggested
solutions for detecting errors and rebuilding PDB
structures are not enough to ensure the correctness of
PDB data before the actual usage since they require a
periodically update (Touw et al., 2014). Yet, the
development of automatic data quality tools is
necessary for checking all the criteria before using to
PDB data.
The Big Data technology is applied to datasets
whose data volume is beyond the ability of commonly
used software tools (i.e., pdb-care program (Lütteke
et al., 2004) and HHsearch (Quignot et al., 2018) to
store, manage, and treat the data within a tolerable
elapsed time. Accordingly, Big Data can support the
expected growth of the PDB database by facing its
challenges (i.e., errors input data) and providing
opportunities to not only store and analyze data but
also allow others to contribute to the improvement of
this database. Thus, it is necessary to involve Big
Data tools in each stage of the management and
process of the PDB database for driving a timely,
accurate and significant knowledge extracted from
the 3D data that represent a value for understanding
the biological phenomena deeply.
Table 2: Data quality issues classified into quality
dimensions.
Data Quality Dimensions
Accuracy
Completeness
Consistency
Missing data
⨉
⨉
Data entry errors
⨉
Incorrect data
⨉
Outdated data
⨉
Contradictory
values
⨉
⨉
Irrelevant data
⨉
Redundancy
⨉
⨉
⨉
5 CONCLUSIONS
In this paper, we have proposed a new model for
processing the big 3D data with specific consideration
of data quality issues. This model has shown how to
position the quality management in (3D) data
analytics. To validate this mode, we have conducted
a case study of 3D PDB data analytics with quality
assessment, where we have firstly summarized all the
possible 3D PDB data quality problems such as
outdated data or contradictory value problems into a
data quality taxonomy, and then classified these the
taxonomy of 3D protein data quality issues into
different data quality dimensions.
As future work, we plan to further standardize the
model in the biological domain, and also validate the
proposed model with different big 3D data in
different domains. We also plan to compare the
similarity and differences of 3D data quality problem
across different application domains.
ACKNOWLEDGEMENTS
The work was supported from European Regional
Development Fund Project "CERIT Scientific Cloud"
(No. CZ.02.1.01/0.0/0.0/16_013/0001802).
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
290

Table 1: An overview of the application of Big Data for detecting errors in Protein data bank Files.
Big Data
tools are
discussed?
No
No
No
Solutions
Propose the pdb
-care program to align the 3D
information with the reported assignments
and support annotation of complex
carbohydrate structures in PDB files.
Ensure the quality of published structures by
validating the new PDB information before
the onlin
e publication, where, a report would
include a brief summary of global quality
indicators, as well as more detailed PDB
information that would allow one to judge
whether specific conclusions are justified by
the quality of the PDB data.
Use the PDB_REDO se
rver to refine and
validate data submitted to PDB platform.
Countermeasure
Cleaning
Filtering
Annotation
Cleaning
Filtering
Cleaning
Filtering
Description of problems
Error detection of 30% of PDB entries (version
September 2003) containing carbohydrates comprise
errors in glycan description.
Detection of gross errors in PDB files (i.e., tracing
the chain backward).
Detection of PDB File errors before online
publication
.
Data quality
problem
Text error
Outdated data
Input error
Outdated data
Input error
References
(Lütteke et al.,
2014)
(Read et al.,
2011)
(Joosten et al.,
2014)
Yes
No
Yes
Implement a peptide
-validation method in
WHAT_CHECK interface to
detect peptide
bonds that need the correction.
Store all the PDB anomalies and errors in
PDBREPORT database.
Enter a PDB accession code in the
PDBREPORT database and get a report of
possible errors in that input file.
Use WHY_NOT database to explain why
PDB entries in any databank do not exist.
Cleaning
Filtering
Classification
Cleaning
Filtering
Storage
Search
Indexation error
Cleaning
Filtering
Storage
Detection of incorrect trans
-cis flips and peptide
-plane
flips in the Protein Data Bank.
Detection of PDB data errors and anomalies.
During the indexation of all PDB entries for each
databank, there are some PDB entries that are missing.
Input error
Contradictory
values
Irrelevant data
Anomalies
Outdated data
Indexation error
(Touw et
al.,
2015)
(Touw et
al., 2014)
(Touw et
al., 2014)
Quality Management for Big 3D Data Analytics: A Case Study of Protein Data Bank
291

Table 1: An overview of the application of Big Data for detecting errors in Protein data bank Files. (cont.)
Big Data tools
are discussed?
No
No
No
No
Solutions
Develop PDBFINDER database as a
searchable PDB entry
meta-data and hold a lot of quality information about the
PDB entries.
Develop new algorithms to allow automatic rebuilding and
remodeling of crystallographic structures in the Protein
Data Bank.
Update PDB_REDO pipeline.
Propose an ap
proach based on semiempirical models for
creating a more realistic working model from a PDB entry
Use the PDB
-REDO pipeline for re
-annotating and re
-
refining the glycoprotein structure models from the Protein
Data Bank.
Countermeasure
Search
Cleaning
Filtering
Search
Cleaning
Filtering
Storage
Search
Cleaning
Filtering
Search
Cleaning
Filtering
Description of problems
Detection of PDB data errors and anomalies.
Difficulty to search in PDB
-format.
Development of tools for
correcting PDB errors prior
to deposition.
Detection of the anomalies in PDB files
Detection of errors in glycoprotein structure models
from the Protein Data Bank.
Data quality
problem
Input error
Search
Input error
Outdated data
Input error
Input error
Contradictory
values
References
(Touw et al.,
2014)
(Joosten et al.,
2011)
(Brandon et
al., 2015)
(Van et al.,
2018)
No
Yes
Yes
Yes
Use PDB
-REDO procedure to re
-refine and rebuild
macromolecular structures before and after they are
submitted to the Protein Data Bank.
Propose PixelDB (Peptide Exosite Location Database) for
searching and modeling protein
–peptide structures
Using clustering for analyzing the structures and limiting
the data redundancy.
Propose PPI3D as a web server for
searching and modeling
pairwise protein
–protein interactions in the context of 3D
structure.
Use PPI3D to reduce the data redundancy by clustering
and analyzing the properties of protein
–protein interactions
Propose InterEvDock2 as a protein docking serve
r for
automatic template search based on HHsearch program and
comparative modeling of the input protein data.
Using clustering to analyze and exclude redundant data
sequences.
Search
Filtering
Analyze
Search
Analyze
Search
Analyze
Search
Optimization of crystallographic structure models in
the PDB by considering the ignored hydrogen bond
(H-bond) distances as a source of information.
Dealing with protein
–peptide complexes
Prediction of structures of protein
–protein interactions
in the cont
ext of 3D structures
Prediction of structures of protein
-protein interactions
without redundant sequences
Using sequences and 3D structures as input data
Search
Filtering
Contradictory
values
Irrelevant
data
Redundancy
Redundancy
Redundancy
(Van et al.,
2018)
(Frappier et
al., 2018)
(Dapkūnas et
al., 2018)
(Quignot, et
al., 2018)
IoTBDS 2019 - 4th International Conference on Internet of Things, Big Data and Security
292

REFERENCES
Abriata, L. A. (2016). Structural database resources for
biological macromolecules. Briefings in
bioinformatics, 18(4), 659-669.
Aydin, Z., Kaynar, O., Görmez, Y., And Işik, Y. E. (2018).
Comparison of machine learning classifiers for protein
secondary structure prediction. 26th Signal Processing
and Communications Applications Conference.
Berman, H. M., Kleywegt, G. J., Nakamura, H., And
Markley, J. L. (2014). The Protein Data Bank archive
as an open data resource. Journal of Computer-aided
Molecular Design, 28(10), 1009-1014.
Brandon, C. J., Martin, B. P., McGee, K. J., Stewart, J. J.,
And Braun-Sand, S. B. (2015). An approach to creating
a more realistic working model from a protein data bank
entry. Journal of molecular modeling, 21(1).
Chen, Z., Zhao, P., Li, F., Leier, A., Marquez-Lago, T. T.,
Wang, Y., and Song, J. (2018). iFeature: a python
package and web server for features extraction and
selection from protein and peptide sequences.
Bioinformatics.
Corral-Corral, R., Chavez, E., And Del Rio, G. (2015).
Machine learnable fold space representation based on
residue cluster classes. Computational Biology and
Chemistry, 59, pp. 1-7.
Correa, L., Borguesan, B., Farfán, C., Inostroza-Ponta, M.,
And Dorn, M. (2018). A Memetic Algorithm for 3D
Protein Structure Prediction Problem. IEEE/ACM
transactions on computational biology and
bioinformatics, 15(3), 690-704.
Crivellaro, A., Rad, M., Verdie, Y., Yi, K. M., Fua, P., And
Lepetit, V. (2018). Robust 3D object tracking from
monocular images using stable parts. IEEE
Transactions on Pattern Analysis and Machine
Intelligence, 40(6), pp. 1465-1479.
Dai, A., Nießner, M., Zollhöfer, M., Izadi, S., And
Theobalt, C. (2017). Bundlefusion: Real-time globally
consistent 3d reconstruction using on-the-fly surface
reintegration. ACM Transactions on Graphics, 36(4).
Dapkūnas, J., Timinskas, A., Olechnovič, K.,
Margelevičius, M., Dičiūnas, R., And Venclovas, Č.
(2017). The PPI3D web server for searching, analyzing
and modeling protein–protein interactions in the
context of 3D structures. Bioinformatics, 33(6).
Frappier, V., Duran, M., And Keating, A. E. (2018).
PixelDB: Protein–peptide complexes annotated with
structural conservation of the peptide binding mode.
Protein Science, 27(1), pp. 276-285.
Ge, M., Helfert, M. (2008), Effects of information quality
on inventory management. International Journal of
Information Quality, 2(2), pp. 177-191.
Ge, M., Helfert, M. (2006), A Framework to Assess
Decision Quality Using Information Quality
Dimensions. 11th International Conference on
Information Quality, pp.455-466.
Helfert, M., Foley, O., Ge, M., Cappiello, C. (2009),
Analysing the effect of security on information quality
dimensions. 17th European Conference on Information
Systems, pp.2785-2797, 2009.
Joosten, R. P., Joosten, K., Cohen, S. X., Vriend, G., And
Perrakis, A. (2011). Automatic rebuilding and
optimization of crystallographic structures in the
Protein Data Bank. Bioinformatics, 27(24).
Joosten, R. P., Long, F., Murshudov, G. N., And Perrakis,
A. (2014). The PDB_REDO server for macromolecular
structure model optimization. International Union of
Crystallography, 1(4), 213-220.
Kalaivani, S., Ramyachitra, D., And Manikandan, P.
(2018). K-means Clustering: An Efficient Algorithm
for Protein Complex Detection. In Progress in
Computing, Analytics and Networking, pp. 449-459.
Li, Q., Li, W., Zhang, J., And Xu, Z. (2018). An improved
k-nearest-neighbor method to diagnose breast cancer.
Analyst.
Lütteke, T., And Von Der Lieth, C. W. (2004). pdb-care
(PDB carbohydrate residue check): a program to
support annotation of complex carbohydrate structures
in PDB files. BMC bioinformatics, 5(1).
Palenzuela, C. L. M., And Pumera, M. (2018). (Bio)
Analytical chemistry enabled by 3D printing: Sensors
and biosensors. TrAC Trends in Analytical Chemistry.
Quignot, C., Rey, J., Yu, J., Tufféry, P., Guerois, R., And
Andreani, J. (2018). InterEvDock2: an expanded server
for protein docking using evolutionary and biological
information from homology models and multimeric
inputs. Nucleic acids research.
Read, R. J., Adams, P. D., Arendall III, W. B., Brunger, A.
T., Emsley, P., Joosten, R. P., and Perrakis, A. (2011).
A new generation of crystallographic validation tools
for the protein data bank. Structure, 19(10), 1395-1412.
Tiwari, A. K., And Srivastava, R. (2018). An Efficient
Approach for Prediction of Nuclear Receptor and Their
Subfamilies Based on Fuzzy k-Nearest Neighbor with
Maximum Relevance Minimum Redundancy. Physical
Sciences, 88(1), pp. 129-136.
Touw, W. G., Joosten, R. P., And Vriend, G. (2015).
Detection of trans–cis flips and peptide-plane flips in
protein structures. Biological Crystallography, 71(8).
Touw, W. G., Baakman, C., Black, J., te Beek, T. A.,
Krieger, E., Joosten, R. P., And Vriend, G. (2014). A
series of PDB-related databanks for everyday needs.
Nucleic acids research, 43(D1), pp. 364-368.
Tripathy, R., Mishra, D., Konkimalla, V. B., And Nayak, R.
K. (2018). A computational approach for mining
cholesterol and their potential target against GPCR
seven helices based on spectral clustering and fuzzy c-
means algorithms. Journal of Intelligent and Fuzzy
Systems, pp. 1-10.
Van Beusekom, B., Lütteke, T., And Joosten, R. P. (2018).
Making glycoproteins a little bit sweeter with PDB-
REDO. Acta Crystallographica Section F: Structural
Biology Communications, 74(8).
Vignesh, U., And Parvathi, R. (2018). 3D visualization and
cluster analysis of unstructured protein sequences using
ARCSA with a file conversion approach. The Journal
of Supercomputing, 1-15.
Wang, R.Y. and Strong, D.M. (1996), Beyond accuracy:
what data quality means to data consumers. Journal of
Management Information Systems, 12(4), pp. 5-34.
Quality Management for Big 3D Data Analytics: A Case Study of Protein Data Bank
293
