
Phylogeny of Kemenyan Toba (Styrax sumatrana) Inferred
from trnl-trnf Chloroplast DNA Sequence
Arida Susilowati
1
, Henti Hendalastuti Rachmat
2
,
Wiza Noni Fadilah
1
and Yosie Syadza Kusuma
1
1
Faculty of Forestry, Universitas Sumatera Utara. Jl. Tri Dharma Ujung No. 1, Kampus USU,
Medan 20155, North Sumatra, Indonesia
2
Forest Research, Development and Innovation Agency Ministry of Environment and Forestry. Jl. Gunung Batu No. 5. PO
Box 165, Bogor 16001, West Java, Indonesia. Tel.: +62-251-8633234; 7520067. Fax. +62-251-8638111
Keywords: DNA, Styrax sumatrana, trnL-trnF, Phylogeny.
Abstract: Styrax sumatrana is a member of genus styrax that cultivated by local comunities in North Sumatra due to its
higher rosin and cinnamic acid content compared to others. This species is widely distributed in Tapanuli
Utara, Pakpak Bharat, and Humbang Hasundutan District. The information on Styrax sumatrana molecular
phylogeny in North Sumatra has not determinet yet, whereas it is important for future breeding and
conservation efforts. Therefore, this research was conducted to determined the phylogenetic relationship
Styrax sumatrana in North Sumatra and other member of genus Styrax in the world. The material for genetic
analysis were leaves sample from 10 individuals and collected from Humbang Hasundutan, Tapanuli Utara,
and Pakpak Bharat. Samples then extracted by using CTAB (Cetyl Trimethyl Ammonium Bromide) method.
DNA amplification was perform using PCR with annealing temperature 50°C. Sequence data analysis was
conducted by using BioEdit software and phylogenetic tree construction was using Mega 5.05. The results
showed that 3 sampled populations of S. sumatrana were grouped into four haplotypes. Phylogenetic tree
analysis result showed that Styrax sumatrana has the closest relationship with Styrax suberifolius and Styrax
chinensis, both are Chinese kemenyan species, with 63% bootstrap value.
1 INTRODUCTION
Indonesia known as megabiodiversity country with
huge number of endemic flora and fauna. Information
on plant species diversity in Indonesia is needed for
future conservation strategy and loosen the rate of
diversity loss. Among those of important native and
multipurpose tree in North Sumatra comes from
Styrax Genus, and locally are known as kemenyan
species. Steenis (1953) mentions four species of
kemenyan which were found and cultivated by local
farmer in North Sumatra, those were: kemenyan toba
(Styrax sumatrana J.J.SM), kemeyan durame (Styrax
benzoin), kemenyan siam (Styrax tonkinennsis) and
kemenyan bulu (Styrax paralleloneurum). There are
several local names standing both for similar and
different species of the Styrax.
Styrax sumatrana known by local people as the
best rosin producer in North Sumatera compared to
other because of it whitish color (preffered by market)
and stronger odor in which they were assumed have
higher rosin and cinnamic acid content (Hidayat et al.
2018). Rosin from this family has long been known
for local medicinal, traditional event and
pharmaceutical purpose and globally named as
benzoin resin (Susilowati et al. 2017).
The general purpose of phylogenetic
reconstruction using molecular evidence is done on
the basis of a homology sequence by aligning DNA
sequences (Thomy et al. 2018). Variations in the
sequence of nucleotide caused by substitution of base
or the indel. Compared to another marker, the
secondary structure of the intron trnL is often
constructed to infer homology position, for example
in Annonaceae (Pirie et al. 2007). Among the plant
DNA regions, non-coding areas such as trnL-trnF and
trnH-psbA chloroplast markers usually exhibit high-
level variations, including indel polymorphism, and
for some cases can provide good capacity for species
identification (Rachmat et al. 2017)
The use of non-coding region sequences of
chloroplast genome also has potential in phylogenetic
research (Soltis and Soltis 1998). Non-coding region
trnL-trnF is region with the highest mutation
frequency so that in most varied trnL-trnF sequence
plants (Taberlet et al. 1991). Therefore, our research
26
Susilowati, A., Rachmat, H., Fadilah, W. and Kusuma, Y.
Phylogeny of Kemenyan Toba (Styrax sumatrana) Inferred from trnL-trnF Chloroplast DNA Sequence.
DOI: 10.5220/0008387300260029
In Proceedings of the International Conference on Natural Resources and Technology (ICONART 2019), pages 26-29
ISBN: 978-989-758-404-6
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
was conducted to determined phylogenetic
relationship of kemenyan toba (Styrax sumatrana) in
North Sumatra. An understanding of genetic identity
of the species and/or population is great practical
importance both to conservation biologists and
silviculturist.
2 MATERIAL AND METHODS
2.1 Plant Material
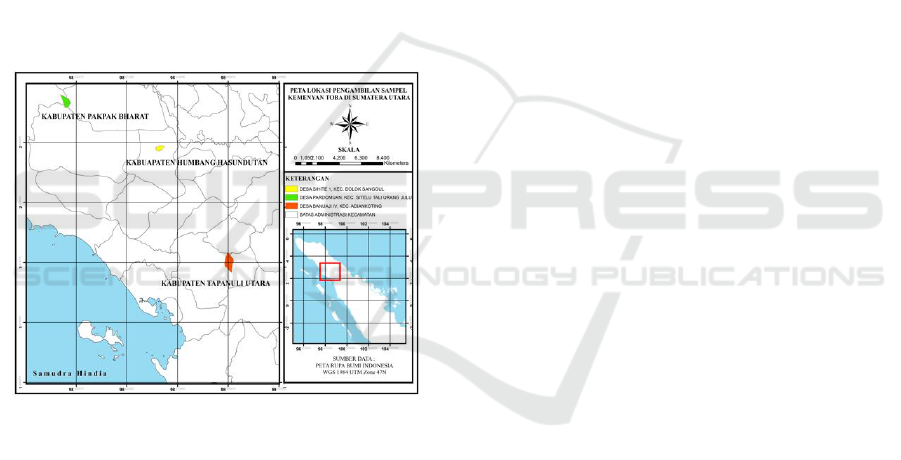
Kemenyan leaf sample collected from tree different
populations those were Humbang Hasundutan,
Pakpak Bharat and North Tapanuli (Figure 1). This
population were choosen based on their different of
altitude and also the presence of physical barrier of
Bukit Barisan mountain range. Ten individuals S.
Sumatrana was taken from healthy and mature trees
of each population.
Figure 1. The origin of kemenyan sample from tree
population.
The total of 30 samples were sampled originated
from 15 years or more mature trees with the average
of dbh was 21 cm. Leaf samples were cut to ± 2 cm x
2 cm size and then stored into an plastic clip filled
with silica gel at a ratio of 1:5, and kept until all leaf
samples ready for extracted. The total genomic DNA
was extracted by using a modified CTAB (Cetyl
Trimethyl Ammonium Bromide) method according
Murray and Thompson (1980).
2.2 DNA Sequencing of trnL-trnF
Chloroplast DNA (cpDNA)
The trnL-trnF cloroplast region was amplified by
PCR using the universal c and f primers described in
Taberlet et al. (1991). PCR process was performed
using 20 uL of a solution containing 10 ng of genomic
DNA, 5 pmol of each forward and backward primer,
and 10 uL of Go Taq® Hot Start Colourless Master
Mix (Promega, Wisconsin, USA) according to the
manufacturer’s instructions.
Initial denaturation was performed at 95°C for 2
min, followed by 30-35 cycles of denaturation at
95°C for 1 min, annealing at 50°C and polymerization
at 72°C for 45 min, and final extension at 72°C for 7
min. Prior to sequencing, the PCR products were
purified and automatic sequenced by Genetic Science
(Singapore). DNA sequencing was performed for
both strands with the primers were used for the PCR
amplifications.
2.3 Data Analysis
The successful rate of amplification then assembled
using a nucleotide assembly software. In this study
the assembly of nucleotides more clearly using
BioEdit software (Hall, 1999). Sequences are aligned
using MEGA 5.05 software (Tamura et al. 2011) in
the ClustalW menu (Larkin et al. 2007) and then
manually adjusted.
Phylogenetic studies were analyzed using MEGA
5.05 software on the phylogeny menu using the
Neighbor-Joining (NJ) method. The consistency of
NJ phylogenetic trees was tested by the bootstrap
method (Felsenstein, 1985) of 1,000 repetitions.
Genetic distance between samples was analyzed
using Kimura 2-parameter method (K2P) (Kimura,
1980). Phylogenetic studies of another Styrax species
using trnL-trnF primers can be obtained from the
sequence databases of various deposited Styrax types
in NCBI (https://www.ncbi.nlm.nih.gov), the DNA
sequence is then aligned with Mega 5.05 using Align
by Muscle and then select the Phylogeny menu to
obtain the phylogenetic tree.
3 RESULT AND DISCUSSION
Not all of 30 samples produce clearly
chromatographic sequences and graph, at the end we
only used 26 individuals that yield clear and unbiased
sequence read. The four excluded individuals were
originated two from Humbang Hasundutan (SS13HB,
and SS20HB) and two more individuals from
Tapanuli Utara (SS23TU and SS24TU).
After alignment we obtained 941 bp of sequence
length from all individuals which were divided into 4
haplotypes (Rachmat et al. 2017). Phylogenetic
analysis was performed using Mega 5 software, using
Phylogeny of Kemenyan Toba (Styrax sumatrana) Inferred from trnL-trnF Chloroplast DNA Sequence
27

Neighbour Joining (NJ) method. The analysis of the
trnL-trnF gene involved 53 data, containing 26
Styrax sumatrana sequences and 27 other Styrax
species, including:. Others styrax sequences
references were downloaded from NCBI. Styrax
suberifolius, Styrax chinensis, Styrax gentryl, Styrax
pentlandianus, Styrax nunezii, Styrax latifolius,
Styrax peruvianus, Styraz camporum, Styrax
leprosus, Styrax pohlii, Styrax obtusifolius, Styrax
ferrugineus, Styrax rotundatus, Styrax acoustic,
Styrax tomentosus, Styrax lanceolatus, Styrax glaber,
Styrax portoricensis, Styrax martii, Styrax laberi,
Styrax ubargenteus, Styrax officinalis, Styrax
benzoin, Styrax aureus, Styrax japonicus, and Styrax
agrestis. The reference sequences were aligned using
the Align by Muscle menu. The phylogenetic tree is a
graph used to describe the interconnecting kinship
between species consisting of a number of nodes and
branches with only one branch connecting the two
closest nodes. Each node represents the taxonomic
units and each branch represents the relationships
between units that describe the hereditary relationship
with the ancestor.
The phylogenetic tree produced by the Neighbour
Joining (NJ) method produces a hypothesis of kinship
relationships between samples based on the genetic
distance in the trnL-trnF gene. In the present study,
phylogenetic trees were tested statistically using the
bootstrap method of 1000 replications presented in
Figure 2.
Reconstruction of phylogenetic trees based on
molecular markers trnL-trnF shows the separation of
several groups. The sample group of Styrax
sumatrana is supported with 83% bootstrap values
consisting of four sub-groups with bootstrap values
ranging from 56 – 94 % (See Figure 2). From the
phylogenetic tree, it can be seen that Styrax
sumatrana has the closest relationship with two China
species of Styrax suberifolius and Styrax chinensis
with a bootstrap value of 63%. Even though shared
similar habitat, the relationship among S. sumatrana
with that of S. benzoin seemed to be far enough.
Considering the phylogenetic relationship as
described in Figure 2, we can determine that S.
sumatrana had close ancestry with those of China
species of Styrax suberifolius and Styrax chinensis
rather than S. benzoin which grow and share similar
habitat type. Phylogenetic trees provide information
about the classification of populations based on their
evolutionary relationships. The roots of the tree
illustrate the first branching point or origin of each
population on the assumption that the rate of
evolution is running constant (Dharmayanti, 2011).
Figure 2: Phylogenetic tree of Styrax sumatrana and the
other Styrax from all over the world with the same marker.
Note : SS (Styrax sumatrana), PB (Pakpak Bharat), TU
(Tapanuli Utara), HB (Humbang Hasundutan).
Among their inter population differences, we
found that the genetic distance between Styrax
sumatrana from Tapanuli Utara and Pakpak Bharat is
0.003 or 99.7% similarity, Tapanuli Utara with
Humbang Hasundutan similarity is 99%, while
Humbang Hasundutan value with Pakpak Bharat is
99.7%.
The genetic distance of Styrax sumatrana with
Styrax chinensis and Styrax suberifolius is 0.005 or
with similarity of 99.5%. While Styrax sumatrana
with Styrax benzoin have similarity of 99,3%.
4 CONCLUSIONS
Our result on the phylogenetic tree of Styrax
sumatrana showed that this species has the same
monophyletic with Styrax suberifolius and Styrax
chinensis with a bootstrap value of 63%. Although S.
sumatrana and S. benzoin were planted together in
North Sumatera, both of species separated into
different group.
ICONART 2019 - International Conference on Natural Resources and Technology
28

REFERENCES
Dharmayanti, I. 2011. Filogenetika molekuler: metode
taksonomi organisme berdasarkan sejarah evolusi.
Wartazoa:21:1
Hall, T.A. 1999. BioEdit: a user-friendly biological
sequence alignment editor and analysis program for
Windows 95/98/NT. Nucl. Acids. Symp. Ser 41: 95-98.
Hidayat, A., Iswanto, A. H., Susilowati, A., Rachmat, H. H.
2018. Radical scavenging activity of Kemenyan Rosin
Produced by an Indonesian native plant, Styrax
sumatrana. Journal of the Korean Wood Sciences
Technology: 46 No.4:346-354
Felsenstein, J. 1985. Confidence limits on phylogenies: an
approach using bootstrap. Evolution 39:783-791.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R.,
McGettigan, P. A., McWilliam, H., Valentin, F.,
Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D.,
Gibson, T. J., Higgins, D. G. 2007. ClustalW and
ClustalX version 2. Bioinformatics 23 (21):2947-2948.
Kimura, M. 1980. A simple methode for estimating
evolutionary rates of base substitutions through
comparative studies of nucleotide sequences. J Mol
Evol 16:111-120.
Murray, H. G., Thompson, W. F. 1980. Rapid isolation of
high molecular weight DNA. Nucleilc Acids Res 8:
4321-4325
Pirie, M. D., Vargas, M. P. B., Bottermans, M., Bakker, F.
T., Chatrou, L. W. 2007. Ancient paralogy in the
cpDNA trnL-F region in Annonaceae. Am J Bot
94:1003-1016.
Rachmat, H. H., Susilowati, A., Elfiati, D., Hartini, K. S,
Faradillah, W. N. 2017. Strong genetic differentiation
of the endemic rosin-producing tree Styrax sumatrana
(Styracaceae) in North Sumatra, Indonesia.
Biodiversitas 18:(4): 1331-1335
Soltis, D. E., Soltis, P. S. 1998. Choosing an approach and
an appropriate gene for phylogenetic analysis. In:
Soltis, D. E., Soltis, P. S., Doyle, J. J., editors.
Molecular Systematics of Plants II: DNA Sequencing.
Netherlands: Kluwer Academic Publishers. ISBN : 0-
412-11131-4. p. 1-42.
Steenis, V. 1953. Styracaceae. Flora Malesiana Ser. I,
Vol.42.
Susilowati, A., Rachmat, H. H., Kholibrina, C. R.,
Ramadhani, R. 2017. Weak delineation of Styrax
species growing in North Sumatra, Indonesia by matK
+ rbcL gene. Biodiversitas 18(3): 1270-1274
Taberlet, P., Gielly, L., Pautou, G., Bouvet, J. 1991.
Universal primers for amplification of three non-coding
regions of chloroplast DNA. Plant Molecular Biology
17:1105-1109.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei,
M., Kumar, S. 2011. MEGA5: Molecular Evolutionary
Genetics Analysis using Maximum Likelihood,
Evolutionary Distance, and Maximum Parsimony
Methods. Molecular Biology and Evolution 28
(10):2731-2739.
Thomy, Z., Yulisma, A., Harnelly, E., Susilowati, A. 2018.
Molecular phylogeny of peat swamp trees in Tripa Peat
Swamp Forest Aceh inferred by 58S Nuclear gene.
Biodiversitas : 19(4):1186-1193
Phylogeny of Kemenyan Toba (Styrax sumatrana) Inferred from trnL-trnF Chloroplast DNA Sequence
29
