
Data-driven Model for Influenza Prediction Incorporating
Environmental Effects
Yosra Didi
1,2 a
, Ahlem Walha
1,2 b
and Ali Wali
2 c
1
Department of Computer Science, Umm Al-Qura University, Makkah, Saudi Arabia
2
Research Laboratory on Intelligent Machines, University of Sfax, National Engineering School of Sfax,
BP1173 Sfax 3038, Tunisia
Keywords:
Prediction, Illness Like Influenza (ILI), LSTM, Machine Learning, Time Series Forecasting, Climatic
Changes, Air Pollution.
Abstract:
Influenza is one of the most severe and prevalent epidemic that causes mortality and morbidity. The researcher
focused on early forecasting to prevent and control the outbreak of the flu disease, which it may reduce
their impact on our daily lives. We propose a model based on machine learning methods that is capable of
making timely influenza prediction using the impact of many environmental factors such as climatic variables,
air pollutants and geographical proximity. Our significant contribution is to incorporate the impact of this
environmental factors changes on the spread of the disease with a machine learning method to improve the
performance of the influenza prediction models. We use multiple data sources including Illness Like Influenza
(ILI) data, climatic factors, air pollutant and geographic proximity that have significant correlation with ILI
rate. In this paper, we compare the proposed model with two methods and with the actual value to prove the
effectiveness of our approach.
1 INTRODUCTION
Influenza is one of the most prevalent and costly dis-
ease that affects many people in the world. Since
2010, Influenza accounts for about 9.2 million to 60.2
million announced diseases in the United States alone
according to the Center for Disease Control (CDC)
1
.
This illness can cause severe health risk and even
death for high level populations. It is contagious
disease resulting in serious respiratory morbidity and
mortality. According to the New York times reports,
the worst influenza season was in 2017-2018.
The current surveillance programs rely on weekly
reports of the data collected from various resources by
health departments on CDC. However, the data col-
lected have a lag time of weeks, therefore, effective
influenza prediction and early outbreak detection are
valuable in surveillance research.
To reduce, prevent and control the outbreak of
influenza in the world, the public health officials
need an effective methods of prediction of influenza
a
https://orcid.org/0000-0003-0845-5731
b
https://orcid.org/0000-0003-2779-5328
c
https://orcid.org/0000-0002-8423-7923
1
https://www.cdc.gov/flu/weekly/index.htm
spread. It will be able to take measures, prioritiz-
ing resources such as emergency rooms, staff and
vaccines. To perform disease surveillance, many re-
searches and studies has been developed. Google Flu
Trends (GFT) was quite successful and used search
engine of Google (Dugas et al., 2013) to predict ILI
activity for more than 25 countries(Ginsberg et al.,
2009) and measure how often a particular term is en-
tered. But it is criticized due to the lack of reliabil-
ity that stopped Google from real time forecasting.
Influenza forecasting research is an active research
area, the existing models have limited ability and ac-
curacy to effectively capture the dynamics of the in-
fluenza spread across different regions. There are sev-
eral methods for time series data analysis based on
machine learning that have earned significant impor-
tance in recent years. In our approach, we focus on
neural network method called Long Short Term Mem-
ory (LSTM) (Hochreiter and Schmidhuber, 1997), it
provides a robust model in temporal data process-
ing. LSTM supersedes Recurrent Neural Network
(RNN)(Siegelmann and Sontag, 1991), it shows re-
markable performance in processing time series data
therefore it attracted much interest in temporal predic-
tion. To improve the accuracy of Influenza forecast-
Didi, Y., Walha, A. and Wali, A.
Data-driven Model for Influenza Prediction Incorporating Environmental Effects.
DOI: 10.5220/0009325500150024
In Proceedings of the 5th International Conference on Internet of Things, Big Data and Security (IoTBDS 2020), pages 15-24
ISBN: 978-989-758-426-8
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
15

ing, we integrated effective external variables that are
shown to have strong influence and impact on flu out-
break. The data explored from different sources are
historical ILI counts, weather information, air pollu-
tion variables, infection status in neighbour regions.
Previous studies such as (Liu et al., 2018) and (Ram
et al., 2015) proved the direct influence of weather
predictable data such as temperature, humidity and
precipitation and for air pollutant like carbon monox-
ide (CO), PM10, Sulphur Dioxide (SO2) and Nitro-
gen Dioxide (NO2) which they are significantly cor-
related with ILI count. Our research objective is to
take advantages of LSTM method, air quality data,
climatic factors and geographical proximity to esti-
mate Influenza in a geographic area within short time
periods (weeks). To this end, we have gathered ILI
data from CDC, pollution censor data from Environ-
mental Protection Agencies (EPA)
2
, climatic variable
and geographical proximity taking from the same ge-
ographic regions and time period to create model for
forecasting Influenza. The experimental results indi-
cate that our approach can predict the influenza trend
very well.
2 LITERATURE REVIEW
Airborne infectious diseases can be extremely expen-
sive, and the more the disease discovery is delayed
the more this cost will increase. The world’s pub-
lic health system has taken considerable attempts to
identify, get ready and control such contagions, and
yet, outbreaks of recent infections, such as the res-
piratory syndrome, persist due to the increasing ur-
banization, but also to the mobility of the contempo-
rary society (Donnelly et al., 2005). So finding an
effective control of airborne infectious disease trans-
mission is still a major issue despite the great work
put in (Luo, 2016). In order to detect influenza in the
early stages, we can set up a public health surveillance
system (or bio-surveillance) whose role is to keep un-
der observation regularly gathered data about patient.
For instance, BioSense (Bradley et al., 2005) is one
of these systems for Disease Control and Prevention:
it was developed by the United States; it gathers pub-
lic health reports from electronic health files in order
to make local and national bio-surveillance an easier
task. Better accuracy of disease detection results in
better reliability of the bio-surveillance system (Wag-
ner et al., 2011). Another concern over performance
is reducing the time lag to notice the outbreak and
retaining large precision of individual disease discov-
2
https://www.epa.gov/outdoor-air-quality-data
ery. In modelling infectious diseases, researchers are
analysing the spreading diseases processes, to predict
the expected outbreak course, and assess the different
strategies deployed to epidemic control.
The prediction research is classified in several cat-
egories. The first category includes Compartmen-
tal models like Susceptible-Infected-Recovered(SIR)
done by (Hethcote, 2000) and (Keeling and Ro-
hani, 2011) and the other model is Susceptible-
Infected-Recovered-Susceptible (SIRS) studied in
(Hooten et al., 2010) and (Shaman et al., 2013)
and Susceptible-Exposed-Infected-Recovered (SEIR)
in (Chowell et al., 2008), (Chowell et al., 2006); in
this models, the population is divided into compart-
ments based on disease states. The movement of in-
dividual between each compartment is defined by a
rate. However, due to the homogeneity of popula-
tion, the model cannot know the patterns with the
variation of age groups and environments. The sec-
ond category is the time series and statistical methods
such as Auto-Regression Integrated Moving Aver-
age (ARIMA) (Choi and Thacker, 1981) and another
method called Generalized Autoregressive Moving
Average (GARMA) (Dugas et al., 2013). These meth-
ods can capture in flexible way the behaviour of in-
fected populations and presume that the values can be
predicted based on past patterns. But their accuracy is
very poor due to the inconsistency of influenza activ-
ity from season to season, especially during the out-
break of diseases. The third category includes meth-
ods of machine learning that became very important
in recent years due to their capability to analyse very
large data called ”Big Data”. There are very popu-
lar machine learning methods such as Sport Vector
Regression(SVR)(Signorini et al., 2011), Neural Net-
work, Binomial Chain (Nishiura, 2011). Opposite
to the statistical methods ARIMA, machine learning
methods considered flexible in the role of capturing
the impact of external factors but expensive in term
of computational, they have to be retrained when new
variable arrives. Recently, special and close attention
was paid to machine learning (ML) classifiers in the
area of influenza detection. In (Elkin et al., 2012)
they had demonstrated how logistic regression clas-
sifier had improved considerably the prediction per-
formance comparing with applying a model to Chief
Complaints from Emergency department.
In the same way, in (Tsui et al., 2011) they had ap-
plied a Bayesian network specific to influenza in order
to analyse this disease in individual patient files; an
expert-constructed tool. (Elkin et al., 2012) and (Tsui
et al., 2011) had chosen the same process: the first
step was to extract the clinical features, then mapped
them to codes using one of the NLP tool, finally,
IoTBDS 2020 - 5th International Conference on Internet of Things, Big Data and Security
16

they used a machine learning method to predict ap-
proximately the risk of epidemic presence. But the
disadvantage is that they take a longer time to de-
tect outbreaks and they didn’t give much attention to
the evaluation of the ML classifiers used. Other than
NLP tools to extract clinical features of influenza, re-
cent work by (Hu et al., 2018) used the data set of
twitter and of the US CDC for influenza reports to
anticipate an approximate real-time improbable per-
centage flu in the United States by region: this was
possible due to the use of an artificial neural network
who owes its success to the improved artificial tree
algorithm (AT)(Li et al., 2017). Search engines and
social networking sites (SNS) are ways faster (7 to
10 days faster) than government organizations (CDC)
in tracking trends of different diseases with real time
analysis on SNS to track ILI (Alessa and Faezipour,
2018). SNS users’ number is increasing exponen-
tially, and people are sharing their daily details in-
cluding their health issues. By consequence, SNS
can provide us with efficient information about health
status; Hence, it can be a useful resource for disease
prediction and control and a better way of communi-
cation to prevent eventual disease outbreaks (Alessa
and Faezipour, 2018). The work done by (Santillana
et al., 2015) apply and test several machine learning
methods on Social Network Twitter. These models
are able to exactly estimate ILI pattern for two weeks
ahead. But, they employed only the features of basic
Bag-of-Word taken from tweets. In (Paul et al., 2016),
the existing NLP techniques has to be enhanced and
developing new methods to effectively explore social
media word and to extract richer Bag-of-Word from
tweets.
The focus of recent studies was on modelling dis-
ease spread in order to analyse the future outbreak
course and assess the potential strategies for disease
mitigation. Besides, the modification of parameters
gives rise to new outcomes; which requires more sce-
nario comparison analysis. For infectious disease
simulations, we need to compare results over space
and time so that we assess decision measures the way
they are implemented with a multitude of state spaces
(Lu et al., 2017).
Analysts need to explore closely the effect of mit-
igated measures; Hence, they need to take advantage
of the environment factors like the effect of changing
weather and the mobility of population.
3 MATERIALS AND METHODS
Our proposed method consists of two steps. In the
first step, we apply a machine learning method which
is the neural network approach LSTM, this technique
is used to predict the initial real time value. In the
next step, we incorporate three different external fac-
tors: (1) Climate variables: temperature, humidity
and precipitation; (2) air pollution sensors includes
different variables such as: PM2.5 concentration, Car-
bon Monoxide (CO) concentration, Nitrogen Diox-
ide (NO2) concentration, PM10 concentration, Sul-
fur Dioxide (SO2) concentration; and (3) geograph-
ical proximity impact is taken from the influence of
neighbouring regions. The objective of integrating the
environmental factors is to reduce the error from the
initial forecast. Figure. 1 shows the architecture fol-
lowed in this paper,
1
CDC ILI Data for
ten regions
Application of
machine
learning
method: LSTM
Data
trained
Air pollution
sensors
Regions
Proximity
Climate Data
External factors
Influenza
Forecasting results
Figure 1: Architecture of the Proposed Model.
3.1 Data Source
Based on previous and recent research, we com-
bine various dataset with LSTM to forecast Influenza
trends. In this research, we focus on CDC-reported
ILI flu counts for ten regions classified by Health and
Human Services (HHS)
3
. The data in reports of CDC
represents the only national dataset with free access
in the United States from 1997-2016. The weather
data is freely accessed and downloaded from Climate
Data Online (CDO)
4
. The climate variables collected
are: Maximum temperature, minimum temperature,
precipitation and humidity. The data of each stations
in the boundaries of the CDC region is aggregated
for each station in each city in each region, by av-
eraging the sum of the time series data into single
weekly. The data of air pollution is collected from the
United States Environmental Protection Agency, the
pollution variables downloaded as mentioned above
are freely accessed. The data is summarized for each
station in each state in each CDC region and calcu-
lated per week. All of the data set collected: flu count,
weather, air pollution variables are pre-processed and
organized by weekly.
3
https://www.hhs.gov/about/agencies/iea/
regional-offices/index.html
4
https:www.ncdc.noaa.gov/cdo-web/datasets
Data-driven Model for Influenza Prediction Incorporating Environmental Effects
17

3.2 Model
The proposed multi-steps approach for Influenza fore-
casting includes the following stages. In the first
stage, we consider a geographical region as a node
for the LSTM approach, this node is trained on the
real ILI counts of regions to predict the original in-
fluenza counts. In the next step, we add the impact of
climate data, geographical proximity and air pollution
sensors to the estimated flu time series after the appli-
cation of LSTM model while the objective is reducing
the error. The LSTM and the proposed approach are
compared with ARIMA model.
3.2.1 Long Short Term Memory Network
The model LSTM is considered as a variation of
RNN architecture. It was designed by Hochreiter and
Schmidhuber (Hochreiter and Schmidhuber, 1997) in
1997, LSTM algorithm is considered faster than the
popular RNN network because it maintains the back-
propagated error in time and layers. LSTM contains
memory blocks which is composed of memory cells
and gates as mentioned in the figure 2. The cell is
performed as a memory while its role is to read, write
and delete information depending on the decisions of
three gates: the gate of input, the gate of output and
the gate of forget. Then each weight of each gate, had
to be trained after the learning process. The memory
cell is implemented as shown in the following equa-
tions: From Eq. (1) to Eq. (5) :
I
t
= σ
g
(W
1
X
t
+U
i
C
t−1
+ b
1
) (1)
F
t
= σ
g
(W
2
X
t
+U
f
C
t−1
+ b
2
) (2)
O
t
= σ
g
(W
3
X
t
+U
o
C
t−1
+ b
3
) (3)
C
t
= F
t
C
t−1
+ I
t
σ
c
(W
c
X
t
+ b
4
) (4)
H
t
= O
t
σ
h
(C
t
) (5)
tanh σ σ
tanh
X X
X
+
σ
H
t-1
x
t
input
Input
gate
Forget
gate
output
gate
S
t-1
H
t
S
t
Figure 2: LSTM Cel Diagram.
Where W and U are the adaptive weights initial-
ized between 0 and 1. F
t
denotes the forget gate vec-
tor, X
t
represents the input vector to the LSTM unit,
It denotes the input gate vector, O
t
is the output gate
vector and H
t
represents the output vector of LSTM
unite. The operator denotes the Hadamard product
(Hochreiter and Schmidhuber, 1997) and (Gers et al.,
1999) and b represents the bias vectors.
The back propagation algorithm is used to train
LSTM cells, where the training criterion is the Mean
square cost function. At the time t − i and to compute
the flu activity O
t−i
, LSTM cell received ILI counts
computed by the previous cell O
t−i−1
and the input
X
t−i
.
The process is repeated for all the LSTM cells in
the model. It is considered that the number of cells
for LSTM is the same number of time steps.
3.2.2 Air Pollution Sensors
The dataset collected contains measures of five types
of pollutants: PM2.5, PM10, NO2, CO, SO2. The in-
fluenza transmission and infection may be due to the
peak of air pollutant concentrations that may play a
significantly important role. We calculated the mean
weekly average for each pollutant, downloaded from
each station in each state belonging to the CDC ten re-
gions. To measure the linear dependence between two
variables, we use the Pearson correlation coefficient.
The value result of this method is between positive 1
which signifies total linear correlation and negative 1
that represents negative correlation, 0 means no linear
correlation. In the Table. 1, we examined the relation-
ship between flu counts and each air pollutant data.
Four pollutant indexes, i.e., CO, NO2, SO2 and PM10
show significant correlation with flu counts. The cor-
relation is significant at the 0.01 level.
Table 1: Correlation Results between Flu Counts and Air
Pollution Data.
CO NO2 SO2 PM10
Pearson correlation 0.95 0.16 0.592 0.361
N 52
The next step, is the decision a priori to investi-
gate the effect of each pollutant on the same week and
lagged by one and two or more weeks as these were
the lags commonly investigated in previous studies.
The dependence of the flu counts on pollutant is very
often with laps of time, that is called lag. To deter-
mine the appropriate lag, we decide to use a crite-
rion like the Akaike Information Criterion (AIC). In
our case, we determine the possible lag impact be-
tween the augmentation of air pollutant variables and
the starting of influenza-like illness, the delay ranging
between 0 and 2 weeks, lag0-lag2: lag0 is considered
as the current week information, lag1 is considered
as the concentration of the previous week and lag2
corresponds to the concentrations of two weeks ear-
lier. The lags between influenza-like illness and each
IoTBDS 2020 - 5th International Conference on Internet of Things, Big Data and Security
18
pollutant concentrations is calculated for all the data
of each region, the total effect Ptot represents the es-
timated of air pollution concentrations for node n at
time t. The following formula Eq. (6) explains the
process.
P
tot
n,t
= Σ
D
i=1
W
n,i
× P
n,i,t
(6)
We used Widrow-Hoff (Widrow and Hoff, 1988)
learning to train the weights, W
n,i
to decrease the
Mean square Error (MSE). D represents the number
of air pollutions sensors and P
n,i,t
denotes the impact
of each pollution variables on the flu counts using the
following formula Eq. (7).
P
n,i,t
=
S
n,t−lag
− S
n,t−lag−1
max(S
n,t−lag
, S
n,t−lag−1
)
(7)
S
n,t−lag
represents the effect estimation at region n
from i
th
air pollution variable at time t. The formula
is the variation before the convenient lag of time and
the actual numeric data P.
3.2.3 Climate Variable Impact
With the objective of including the environmental fac-
tors as input information to the method, first we look
for the relation between the flu counts and the climatic
variables in term of correlation. In the previous stud-
ies, the literature (Soebiyanto et al., 2010), (Lowen
et al., 2007) and (Lowen and Steel, 2014) prove
a strong cross-correlation between minimum, max-
imum temperature and influenza counts from CDC.
Influenza epidemics is often associated with seasonal
changes in temperature and relative humidity. The in-
tegration of different linear time series values is not
an effective method to determine the impact of mete-
orological variables due to the impact delay of tem-
poral variables. According to the study of (Venna
et al., 2019), they compute a situational time lags
between flu counts and each climatic variables, the
daily climatic variable data: temperature, relative hu-
midity and precipitation from CDO, were converted
into values per week by calculating the average. And
each time series were converted to a tuple XY of
symbols: X represents the value magnitude (high,
medium, low) and Y represents the change of value
from the previous time step (increasing, stable, de-
creasing). After the generation of the symbolic tuples
for each value in time series, we compute the frequent
associations between a climate symbolic time series
and Flu symbolic time series at different time lags
from 0 to 5 using the Apriori algorithm [34]. When
the time lags between influenza times series and each
weather predictors are calculated, we calculate the ra-
tio C
n,i,t
of change between the appropriate time lags
and the actual value as explained in the Eq. (8).
C
n,i,t
=
X
n,t−lag
− X
n,t−lag−1
max(X
n,t−lag
, X
n,t−lag−1
)
(8)
Then the total impact is calculated following the
Eq. (9) :
C
tot
n,t
= Σ
D
i=1
W
n,i
× C
n,i,t
(9)
Where t denotes the time steps, n denotes the re-
gion, D the number of climatic variables and W the
weights trained using Widrow-Hoff learning (Widrow
and Hoff, 1988).
(a)
0 10 20 30 40 50 60
Week
0
0.5
1
1.5
2
2.5
Flu Counts
10
4
Region 1
Region 2
Region 3
Region 4
Region 5
(b)
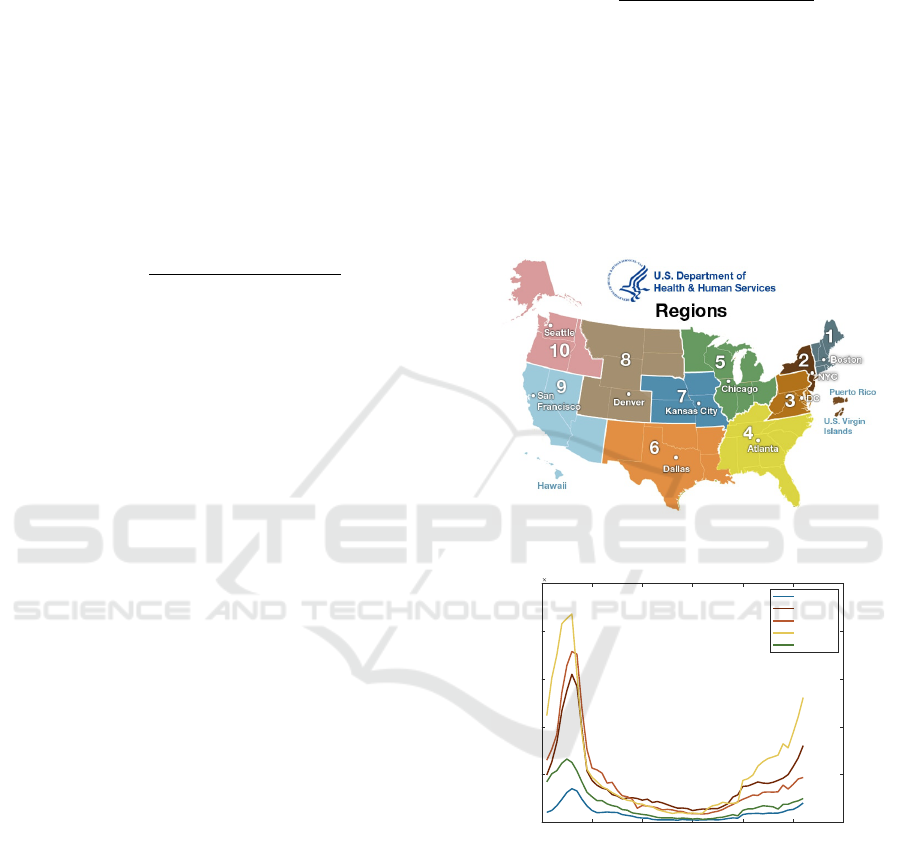
Figure 3: Flu Counts: (a) Division Map of CDC-HHS Re-
gions Taken from HHS Website. (B) a Plot of the Trends
for ILI Counts in 2018 for Five CDC Regions.
3.2.4 Proximity Regions Factor
The geographical proximity has a strong threat of
widespread influenza outbreak. We can observe, as
shown in Fig. 3, a flu trends between regions in spa-
tial proximity. We can calculate the proximity impact
on the node n by a factor coming from each neighbour
nodes which is the average of flu divergence. The fac-
tor A applied at each node n, represents the average
Data-driven Model for Influenza Prediction Incorporating Environmental Effects
19

variation in the flu counts given by the neighbour data
at time t − i. Equation. (10) is as follow:
A
n,i,t
=
1
y
Σ
y
j=1
(F
i,t− j
− F
i,t− j−1
) (10)
An, i, t represents the individual adjustment for the
neighbour i to the node n at time t, is computed with
the ratio change in the previous y steps.
F
i,t− j
denotes the current ILI count for the neigh-
bour i at time t − j. In the study done by (Venna et al.,
2019) the y selected has to be 3, because it gave an op-
timal results. Once the individual adjustment is com-
puted, the total adjustment A
tot
n,t
at time t and node n
is the summation of weights of the individual adjust-
ment A
n,i,t
. The weights are trained using Widrow-
Hoff algorithm (Widrow and Hoff, 1988).
A
tot
n,t
= Σ
N
i=1
W
n,i
× A
n,i,t
(11)
N denotes the number of neighbours of data node
n.
3.2.5 Predict Value Estimation
Final forecast value is calculated after applying the
climate variable impact from Eq. (9), the total ad-
justment factor from Eq. (11) and Eq. (6) for the air
pollution sensors to the predicted value generated by
LSTM at time t for node n as shown in Eq. (12).
F
f inal
n,t
= P
tot
n,t
+C
tot
n,t
+ A
tot
n,t
+ F
LST M
n,t
(12)
4 RESULTS
In this study, we employed ARIMA a time
series-based model compared to the three pro-
posed data-driven models LSTM, proposed model:
LSTM+PS+CI+SA and ARIMA+External factors
(PS+CI+SA) on a freely available data sets about flu
counts from the CDC.
We evaluate our approach on a multiple time se-
ries data: For influenza activity, we download flu
counts real data sets from CDC for all ten HHS re-
gions, the data is weekly presented. We collected 52
weeks of data in ten regions from 1st week of 2018 to
the 52 nd week of 2018, the data selected for training
is from the 1st week to 46 th week. The climate data is
collected from (CDO) which is freely accessed. The
data downloaded from the CDO is weekly aggregated
for each region.
For the air pollution sensors, the data is down-
loaded from the EPA and is freely accessed, than it
was aggregated and averaged into single week.
The external predictors are weekly summarized
time series. Our samples are between 2016-2018, the
training data selected was on 80
4.1 Evaluation Criteria
To evaluate our approach for prediction performance,
we use the following evaluation metrics. Mean Ab-
solute Percentage Error (MAPE): The metric used to
measure the accuracy in percentage, is the average ab-
solute error between actual and estimated values.
MAPE =
1
N
Σ
|A − P|
|A|
(13)
Root Mean Square Percentage Error (RMSPE):
The metric used to compute the deviation between ac-
tual and predicted value and their square root.
RMSPE =
r
1
N
Σ(A − P)
2
(14)
Root Mean Square Error (RMSE): the metric used
to compute the difference between the real value and
predicted value .
RMSE =
s
1
N
Σ
(A − P)
2
A
× 100 (15)
N denotes the number of weeks, A denotes the ac-
tual influenza data and P denotes the predicted value.
As mentioned before, we compared the results of our
approach with the state of art ARIMA method. Also
we compared our results with the predicted value gen-
erated after application of the LSTM to prove that our
method reduces the error after LSTM.
4.2 Results for the CDC Dataset
Table. 2 shows the comparison of three models of pre-
diction: LSTM, ARIMA and our proposed approach
(LSTM + external predictors). These models are ap-
plied on all the geographical HHS regions. The table
compares the forecasting performance of the models
until 25 weeks in the future. We do three experiments
on CDC dataset for the ten regions: one for LSTM,
second for ARIMA and the third experiments for the
proposed model. We computed the performance in
terms of MAPE, RMSPE and RMSE. We can observe
an improvement of prediction accuracy after integrat-
ing the external predictors: pollution sensors, climatic
data and geographical components, to LSTM data, the
error was reduced that shows the importance of their
impact. The improvement in forecasting is noticed
from week 5 to week 15 ahead, while the first week
IoTBDS 2020 - 5th International Conference on Internet of Things, Big Data and Security
20

0 5 10 15 20 25
Week
0
5
10
15
20
25
30
35
MAPE
ILI Region3
Proposed Method
LSTM
ARIMA
ARIMA+External factors
(a)
0 5 10 15 20 25
Week
0
10
20
30
40
50
60
70
MAPE
ILI Region5
Proposed Method
LSTM [16]
ARIMA [7]
ARIMA+External factors
(b)
0 5 10 15 20 25
Week
5
10
15
20
25
30
35
40
45
50
55
RMSPE
ILI Region3
Proposed Method
LSTM
ARIMA
ARIMA+External factors
(c)
0 5 10 15 20 25
Week
10
20
30
40
50
60
70
80
90
100
RMSPE
ILI Region5
Proposed Method
LSTM [16]
ARIMA [7]
ARIMA+External factors
(d)
0 5 10 15 20 25
Week
0
500
1000
1500
2000
2500
RMSE
ILI Region3
Proposed Method
LSTM
ARIMA
ARIMA+External factors
(e)
0 5 10 15 20 25
Week
400
600
800
1000
1200
1400
1600
RMSE
ILI Region5
Proposed Method
LSTM [16]
ARIMA [7]
ARIMA+External factors
(f)
Figure 4: Comparison Using Error Metrics: MAPE (a) and (B), RMSPE (C) and (D), RMSE (E) and (F) between Region 3
(Left) and Region 5 (Right) of the Flu Prediction Models over 25 Weeks Ahead.
ahead predicted doesn’t show any significant amelio-
ration.
Fig. 4 shows the six error charts metrics including
MAPE, RMSPE and RMSE for two regions: Region
3 and Region 5. Table. 2 is correlated with the plots
in Fig. 4 in numbers for Region 5. We can notice that
for the Region 3 for the future prediction weeks, the
MAPE error for ARIMA is less than the other mod-
els between the fourth week and the seventh week.
And our proposed model performs slightly better than
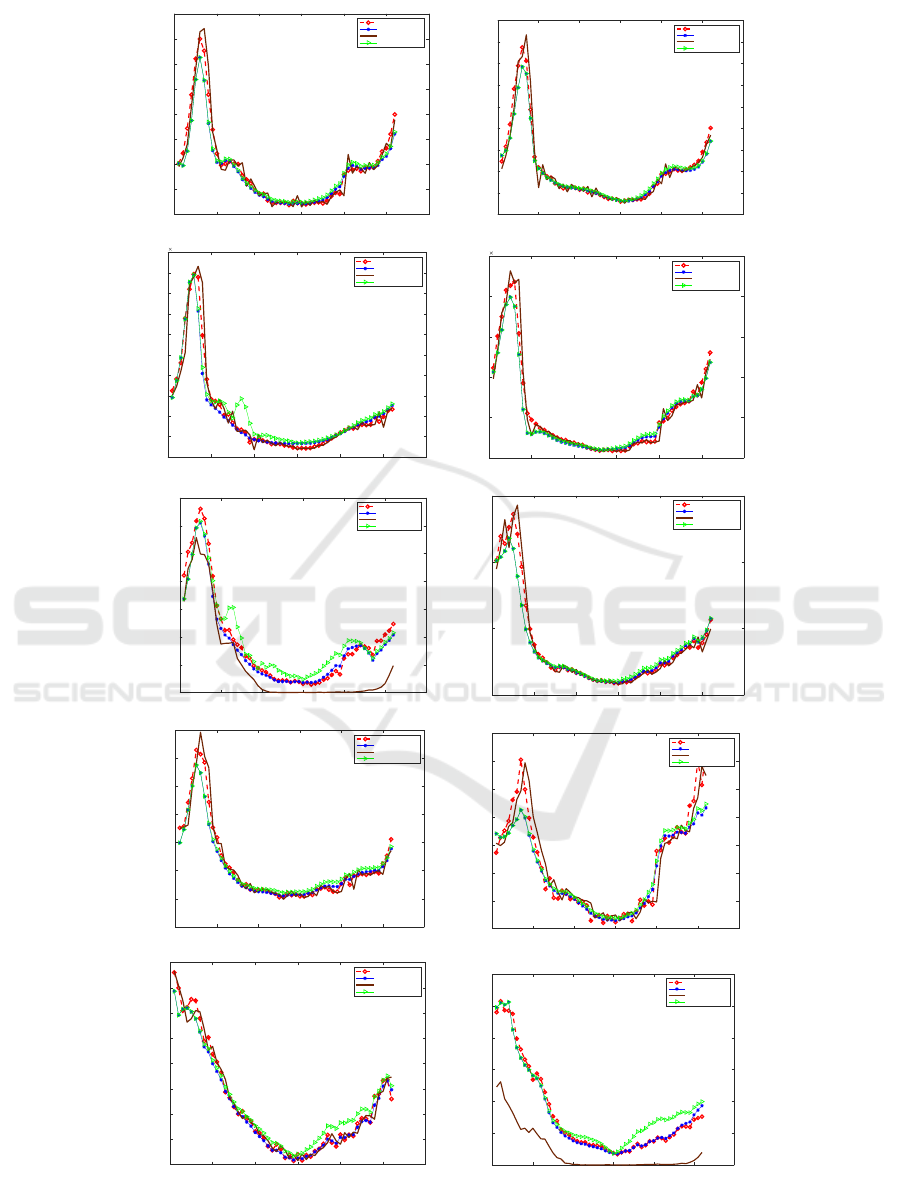
LSTM for the same region. As shown in Fig. 5, there
are ten plots, presenting the ten regions from Region
1 to Region 10 respectively, of the forecasting models
resulting from LSTM method, ARIMA and the pro-
posed compared to the actual value for the year 2018.
It can be seen that the value predicted for all the
regions of the year gives values very close to the ac-
tual one. The approach anticipates an approximate
Data-driven Model for Influenza Prediction Incorporating Environmental Effects
21
0 10 20 30 40 50 60
Week
0
500
1000
1500
2000
2500
3000
3500
4000
Flu Count
ILI Region1
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
2000
4000
6000
8000
10000
12000
14000
16000
18000
Flu Count
ILI Region2
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
Flu Count
10
4
ILI Region3
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
0.5
1
1.5
2
2.5
Flu Count
10
4
ILI Region4
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
1000
2000
3000
4000
5000
6000
7000
Flu Count
ILI Region5
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
5000
10000
15000
Flu Count
ILI Region6
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
-500
0
500
1000
1500
2000
2500
3000
Flu Count
ILI Region7
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
500
1000
1500
2000
2500
3000
3500
Flu Count
ILI Region8
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
500
1000
1500
2000
2500
3000
3500
4000
4500
Flu Count
ILI Region9
Actual
LSTM
ARIMA
Proposed Method
0 10 20 30 40 50 60
Week
0
500
1000
1500
2000
2500
3000
Flu Count
ILI Region10
Actual
LSTM
ARIMA
Proposed Method
Figure 5: A Plots Represent Actual and Predicted Flu Count for 10 Regions.
IoTBDS 2020 - 5th International Conference on Internet of Things, Big Data and Security
22
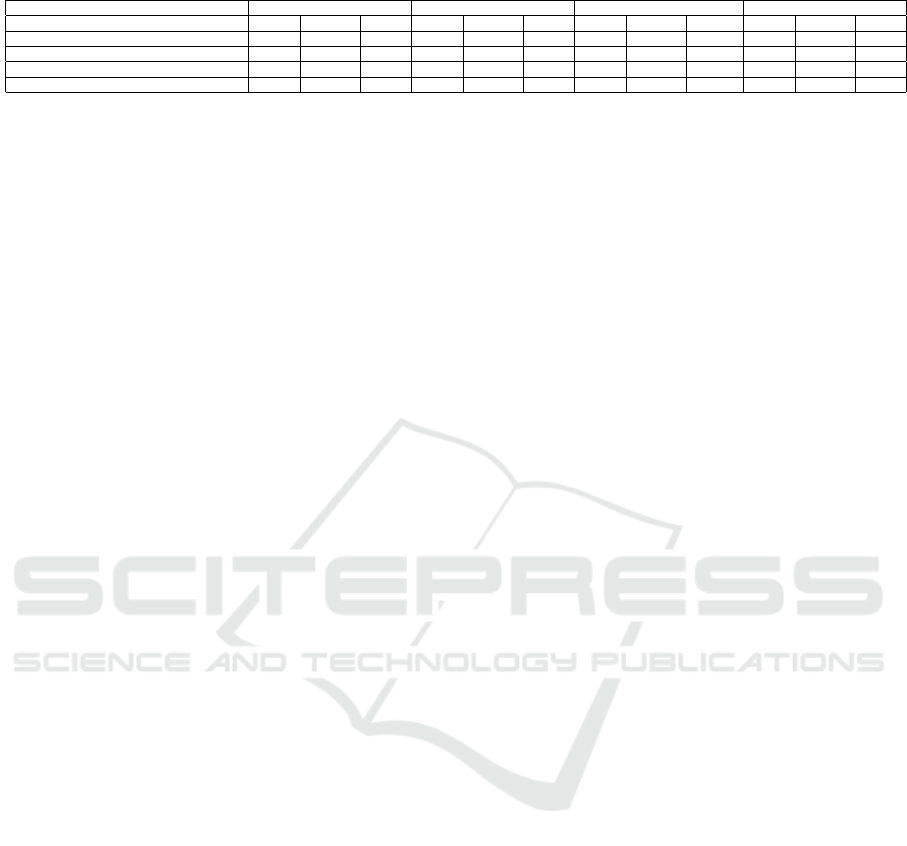
Table 2: MAPE, RMSPE and RMSE for the ILI Count Predicted Using ARIMA and Proposed Model of Region 5.
Weeks 1-week 5-week 10-week 15-week
Model MAPE RMSPE RMSE MAPE RMSPE RMSE MAPE RMSPE RMSE MAPE RMSPE RMSE
LSTM (Hochreiter and Schmidhuber, 1997) 31.95 31.95 1364 21.15 22.59 1134.9 23.92 25.02 1113.35 28.42 29.92 998.89
Proposed model 20.09 20.09 846.8 11.59 13.42 650.18 8.18 10.42 520.37 17.04 23.41 599.73
ARIMA (Choi and Thacker, 1981) 22.23 22.23 937 15.87 17.05 957.68 18.13 19.58 912.08 19.68 21.62 791.33
ARIMA+External factors 22.23 22.23 937 16.03 17.30 977.41 13.88 15.97 850.58 22.07 30.01 845.77
real-time data better than the other model. Accord-
ing to these three errors in Fig. 4 and Fig. 5, we can
say that the proposed approach is favourable for the
prediction of influenza-Like illness.
5 CONCLUSIONS
In this study, we proposed an approach to enhance in-
fluenza prediction. In our contribution, first step is the
application of LSTM as machine learning technique,
this method shows a better performance comparing
to the existing time series prediction methods. Sec-
ond step is the integration of the impacts of the exter-
nal predictors : air pollution data, climatic variables
and geographical proximity whose goal is to reduce
the error of machine learning method. We evaluated
the approach we proposed on the datasets from CDC-
HHS ILI. The proposed approach is compared with
ARIMA model. It can be seen that with the integra-
tion of external predictors in LSTM, we improved the
accuracy performance. Also, the proposed approach
may be useful for other viral illness such as Asthma,
Chickenpox and Ebola. Our future study seeks to
implement the proposed approach on Social Network
Site like Twitter and Instagram dataset.
REFERENCES
Alessa, A. and Faezipour, M. (2018). A review of in-
fluenza detection and prediction through social net-
working sites. Theoretical Biology and Medical Mod-
elling, 15(1):2.
Bradley, C. A., Rolka, H., Walker, D., and Loonsk, J.
(2005). Biosense: implementation of a national
early event detection and situational awareness sys-
tem. MMWR Morb Mortal Wkly Rep, 54(Suppl):11–
19.
Choi, K. and Thacker, S. B. (1981). An evaluation
of influenza mortality surveillance, 1962–1979: I.
time series forecasts of expected pneumonia and in-
fluenza deaths. American journal of epidemiology,
113(3):215–226.
Chowell, G., Miller, M., and Viboud, C. (2008). Seasonal
influenza in the united states, france, and australia:
transmission and prospects for control. Epidemiology
& Infection, 136(6):852–864.
Chowell, G., Nishiura, H., and Bettencourt, L. M. (2006).
Comparative estimation of the reproduction num-
ber for pandemic influenza from daily case notifica-
tion data. Journal of the Royal Society Interface,
4(12):155–166.
Donnelly, C. F., Riley, S., Ferguson, N. M., and Anderson,
R. M. (2005). Transmission dynamics and control of
the viral aetiological agent of sars. Severe Acute Res-
piratory Syndrome: A Clinical Guide, page 111.
Dugas, A. F., Jalalpour, M., Gel, Y., Levin, S., Torcaso, F.,
Igusa, T., and Rothman, R. E. (2013). Influenza fore-
casting with google flu trends. PloS one, 8(2):e56176.
Elkin, P. L., Froehling, D., Wahner-Roedler, D., Brown,
S. H., and Bailey, K. R. (2012). Comparison of nat-
ural language processing biosurveillance methods for
identifying influenza from encounter notes. Annals of
internal medicine, 156 1 Pt 1:11–8.
Gers, F. A., Schmidhuber, J., and Cummins, F. (1999).
Learning to forget: Continual prediction with lstm.
Ginsberg, J., Mohebbi, M. H., Patel, R. S., Brammer, L.,
Smolinski, M. S., and Brilliant, L. (2009). Detecting
influenza epidemics using search engine query data.
Nature, 457(7232):1012.
Hethcote, H. W. (2000). The mathematics of infectious dis-
eases. SIAM review, 42(4):599–653.
Hochreiter, S. and Schmidhuber, J. (1997). Long short-term
memory. Neural computation, 9(8):1735–1780.
Hooten, M. B., Anderson, J., and Waller, L. A. (2010). As-
sessing north american influenza dynamics with a sta-
tistical sirs model. Spatial and spatio-temporal epi-
demiology, 1(2-3):177–185.
Hu, H., Wang, H., Wang, F., Langley, D., Avram, A., and
Liu, M. (2018). Prediction of influenza-like illness
based on the improved artificial tree algorithm and ar-
tificial neural network. Scientific reports, 8(1):4895.
Keeling, M. J. and Rohani, P. (2011). Modeling infectious
diseases in humans and animals. Princeton University
Press.
Li, Q., Song, K., He, Z., Li, E., Cheng, A., and Chen, T.
(2017). The artificial tree (at) algorithm. Engineering
Applications of Artificial Intelligence, 65:99–110.
Liu, L., Han, M., Zhou, Y., and Wang, Y. (2018). Lstm
recurrent neural networks for influenza trends predic-
tion. In International Symposium on Bioinformatics
Research and Applications, pages 259–264. Springer.
Lowen, A. C., Mubareka, S., Steel, J., and Palese, P.
(2007). Influenza virus transmission is dependent on
relative humidity and temperature. PLoS pathogens,
3(10):e151.
Lowen, A. C. and Steel, J. (2014). Roles of humidity and
temperature in shaping influenza seasonality. Journal
of virology, 88(14):7692–7695.
Data-driven Model for Influenza Prediction Incorporating Environmental Effects
23

Lu, Y., Garcia, R., Hansen, B., Gleicher, M., and Maciejew-
ski, R. (2017). The state-of-the-art in predictive visual
analytics. In Computer Graphics Forum, volume 36,
pages 539–562. Wiley Online Library.
Luo, W. (2016). Visual analytics of geo-social interaction
patterns for epidemic control. International journal of
health geographics, 15(1):28.
Nishiura, H. (2011). Real-time forecasting of an epidemic
using a discrete time stochastic model: a case study
of pandemic influenza (h1n1-2009). Biomedical engi-
neering online, 10(1):15.
Paul, M. J., Sarker, A., Brownstein, J. S., Nikfarjam, A.,
Scotch, M., Smith, K. L., and Gonzalez, G. (2016).
Social media mining for public health monitoring and
surveillance. In Biocomputing 2016: Proceedings of
the Pacific symposium, pages 468–479. World Scien-
tific.
Ram, S., Zhang, W., Williams, M., and Pengetnze, Y.
(2015). Predicting asthma-related emergency depart-
ment visits using big data. IEEE journal of biomedical
and health informatics, 19(4):1216–1223.
Santillana, M., Nguyen, A. T., Dredze, M., Paul, M. J.,
Nsoesie, E. O., and Brownstein, J. S. (2015). Combin-
ing search, social media, and traditional data sources
to improve influenza surveillance. PLoS computa-
tional biology, 11(10):e1004513.
Shaman, J., Karspeck, A., Yang, W., Tamerius, J., and Lip-
sitch, M. (2013). Real-time influenza forecasts dur-
ing the 2012–2013 season. Nature communications,
4:2837.
Siegelmann, H. T. and Sontag, E. D. (1991). Turing com-
putability with neural nets.
Signorini, A., Segre, A. M., and Polgreen, P. M. (2011).
The use of twitter to track levels of disease activity
and public concern in the u.s. during the influenza a
h1n1 pandemic. PLOS ONE, 6:1–10.
Soebiyanto, R. P., Adimi, F., and Kiang, R. K. (2010).
Modeling and predicting seasonal influenza transmis-
sion in warm regions using climatological parameters.
PloS one, 5(3):e9450.
Tsui, F., Wagner, M., Cooper, G., Que, J., Harkema, H.,
Dowling, J., Sriburadej, T., Li, Q., Espino, J. U., and
Voorhees, R. (2011). Probabilistic case detection for
disease surveillance using data in electronic medical
records. Online journal of public health informatics,
3(3).
Venna, S. R., Tavanaei, A., Gottumukkala, R. N., Raghavan,
V. V., Maida, A. S., and Nichols, S. (2019). A novel
data-driven model for real-time influenza forecasting.
IEEE Access, 7:7691–7701.
Wagner, M. M., Moore, A. W., and Aryel, R. M. (2011).
Handbook of biosurveillance. Elsevier.
Widrow, B. and Hoff, M. E. (1988). Adaptive switching
circuits.
IoTBDS 2020 - 5th International Conference on Internet of Things, Big Data and Security
24
