
Nematode Identification using Artificial Neural Networks
Jason Uhlemann
1
, Oisin Cawley
1
and Thomais Kakouli-Duarte
2
1
gameCORE, Department of Computing, Institute of Technology Carlow, Kilkenny Road, Carlow, Ireland
2
enviroCORE, Department of Science and Health, Institute of Technology Carlow, Kilkenny Road, Carlow, Ireland
Keywords: Convolutional Neural Networks, Image Classification.
Abstract: Nematodes are microscopic, worm-like organisms with applications in monitoring the environment for
potential ecosystem damage or recovery. Nematodes are an extremely abundant and diverse organism, with
millions of different species estimated to exist. This trait leads to the task of identifying nematodes, at a species
level, being complicated and time-consuming. Their morphological identification process is fundamentally
one of pattern matching, using sketches in a standard taxonomic key as a comparison to the nematode image
under a microscope. As Deep Learning has shown vast improvements, in particular, for image classification,
we explore the effectiveness of Nematode Identification using Convolutional Neural Networks. We also seek
to discover the optimal training process and hyper-parameters for our specific context.
1 INTRODUCTION
Convolutional Neural Networks (CNNs) are state-of-
the-art algorithms that have made significant
advances in computer vision tasks, especially in
Image Classification. The CNN processes multi-
dimensional, grid-like forms of data, such as images
and video (LeCun et al., 2015; Goodfellow et al.,
2016). CNNs are a type of neural network which first
attracted attention when used to solve the task of
recognising handwritten digits using the LeNet-5
architecture (LeCun et al., 1998). Since then, the
accuracy of these types of networks has been steadily
increasing, with the most recent EfficientNet models
(Tan and Le, 2019) achieving a Top-5 accuracy on the
ImageNet dataset of 97.1%.
The availability of Deep Learning frameworks,
such as TensorFlow and PyTorch, has made Deep
Learning more accessible to a broader group of people
allowing applications of Deep Learning in many areas,
from healthcare to self-driving cars (Dargan et al.,
2019). Thus, other contexts which require some form
of image recognition should be able to capitalise on this
new capability. One potential use-case is that of
Nematode Identification.
Nematodes have been proven to be good
environmental bioindicators (Wilson and Kakouli-
Duarte, 2009). A bioindicator is a species that can
provide useful information about the status of the
environment. To be classified as a bioindicator, the
species must play an essential role in the ecosystem, be
abundant and not be capable of being killed by low
levels of pollutants (Cortet et al., 1999). Their
responses should also be measurable and reproducible.
However, Nematodes are an incredibly diverse and
abundant group of organisms that live in terrestrial and
aquatic environments (Dodds and Whiles, 2010;
Poinar, 2016). There are many different families of
nematodes, categorised by their feeding behaviours.
These different groups, known as trophic groups,
consist of bacteria feeders, fungi feeders, predators,
omnivores and herbivores (Kennedy and Luna, 2005).
Due to their small size, and with such a variety of
species, it becomes challenging to identify which types
are present in a particular sample. The current method
is a manual one which is time-consuming and
susceptible to error.
In this research, we explore the feasibility of
designing a CNN suitable for classifying microscope
photographs of nematodes. We use entomopathogenic
nematodes (EPN) which are parasitic nematodes that
cause harm to insects by infecting them with insect-
pathogenic bacteria, to allow us to culture batches of
nematodes in the lab. EPNs have been explored for
their potential to replace the use of chemical pesticides,
which can cause contamination in the environment
(Dillman and Sternberg, 2012; de Oliveira et al., 2016).
For this research, we use three different species of
EPNs: Heterorhabditis bacteriophora, Steinernema
carpocapsae and Steinernema feltiae.
Uhlemann, J., Cawley, O. and Kakouli-Duarte, T.
Nematode Identification using Artificial Neural Networks.
DOI: 10.5220/0009776600130022
In Proceedings of the 1st International Conference on Deep Learning Theory and Applications (DeLTA 2020), pages 13-22
ISBN: 978-989-758-441-1
Copyright
c
2020 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
13

While we confine ourselves to specific nematodes
in this research, the automatic feature extraction of a
CNN will prove invaluable to improving the state-of-
the-art in identifying nematodes. The automatic feature
extraction will allow for the possibility of the model
scaling to more groups of nematodes in the future, and
with greater ease than the traditional approaches.
2 EXISTING APPROACHES TO
NEMATODE IDENTIFICATION
Nematode identification can be achieved using their
morphological, biochemical and molecular features
(Seesao et al., 2017). While there are many
approaches, morphological identification is the
cheapest method and is readily available as only a
light microscope is required. The identification
process involves observing the illuminated nematode
under a microscope and referencing a printed
taxonomic key with sketches for comparison. This
process not only is time-consuming but requires a
certain level of expertise in identifying the essential
characteristics of the nematodes themselves. Also,
printed aids can be seen to be too rigid and unreliable
(Bouket, 2012).
2.1 Computer-Aided Approaches
Using technology to aid in nematode identification
goes back to the 1980s. An overview, presented by
(Diederich et al., 2000), shows many different
approaches to computer identification aids. These
approaches include using cluster analysis, similarity
coefficient and expert systems. However, despite
some success, these approaches do not appear to have
been adopted by the wider community (Diederich et
al., 2000).
An attempt to improve the system of the printed
taxonomic key led to the creation of a web application,
NEMIDSOFT, using the genus Merlinius, plant-
parasitic nematodes (Bouket, 2012). NEMIDSOFT
allows the user to enter in morphological
measurements that get compared to a database for the
closest match. With the use of a database, this
application offers more flexibility to be updated and
scaled to other species. It is unclear how well this
application performed, as the website hosting it no
longer exists.
More recently, artificial neural networks have been
investigated as a means to learn specific photographic
features. These algorithms have been successfully
deployed to detect and count the eggs of soybean cyst
nematodes using a convolutional autoencoder
(Akintayo et al., 2016, 2018; Kalwa et al., 2019). Other
examples include identifying different strains of the
nematode species Caenorhabditis elegans based on
video recordings of their behaviour and movements
(Javer et al., 2018). This method showed improvement
over the state-of-the-art with manual-crafted features.
3 METHODOLOGY
In this section, we will describe the overall study
design, including our considerations around the CNN
architecture, training data and parameter tuning.
3.1 Overview
Our high-level design included developing a CNN,
obtaining photographs of nematodes, training the
CNN on these photographs and validating the
resulting network. One of the primary considerations
with any artificial neural network implementation is
what the network structure will be. Given the success
of CNNs in image recognition, we looked to existing
state-of-the-art architectures to choose the most
appropriate.
A second consideration was the training data. For
our specific context, this turned out to be more
problematic, given the differences between juvenile
and adult nematodes. Also, given the lack of existing
photographs of our species, we would need to generate
our images. This requirement raised the concern that
the amount of training and validation data could be
relatively small.
3.2 CNN Architecture
We initially investigated designing a CNN model
from scratch for this task, using inspiration from the
state-of-the-art models. However, designing a neural
network architecture is a time-consuming task and
requires experience and expertise. Therefore, we
decided to use a state-of-the-art model architecture
already available.
As we are using the Keras deep learning library,
there are 13 CNN architectures available to be used,
with or without pre-trained weights. These
architectures include Xception, VGG16, VGG19,
ResNet, Inception, InceptionResNet, MobileNet,
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
14
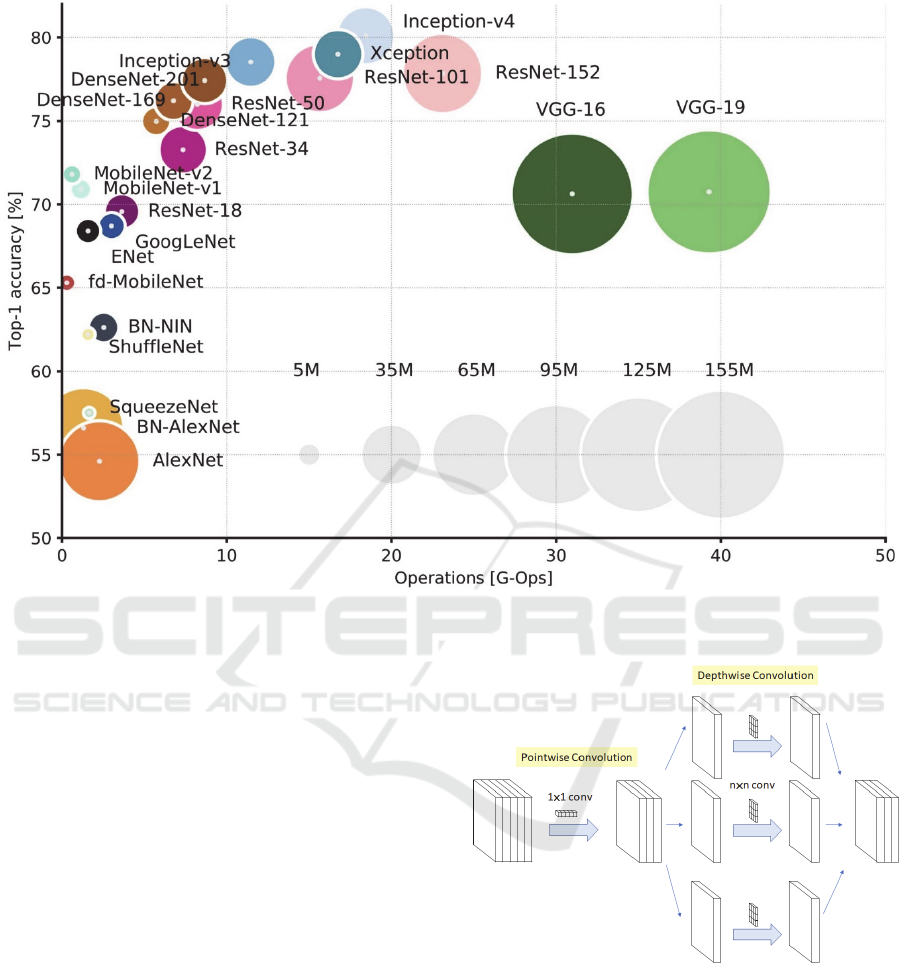
Figure 1: Overview of state-of-the-art models (Culurciello, 2018).
DenseNet and NASNet. To determine which model we
use, we test each model on the same task. This task
consists of training each model for 200 epochs on one
of our generated datasets, without any pre-trained
weights and using the SGD optimiser. The best epoch
for each model is recorded based on the lowest
validation loss value achieved.
Out of the 13 CNN architectures, DenseNet169,
DenseNet201 and NASNetLarge could not train due to
memory limitations. NASNetMobile, VGG16,
VGG19 and MobileNetV2 achieved validation
accuracies between 30% and 55%. This leaves
InceptionResNetV2, DenseNet121, Xception,
ResNet50, InceptionV3 and MobileNet achieving
accuracies between 90% and 100%. Out of all these
models, InceptionResNetV2 has the highest Top-5
accuracy on the ImageNet dataset at 95.3%. However,
InceptionResNetV2 is quite a large model, with a depth
of 572 layers and 55,873,736 parameters. To avoid
potential problems, we decide to use the next best
model, the Xception model (Figure 2), which has a
Top-5 accuracy of 94.5%, a depth of 126 layers and
22,910,480 parameters.
Figure 2: Image of Depthwise Separable Convolutions used
in Xception (Tsang, 2018).
3.3 Data Gathering
To gather the images required, we first culture the
three different species by infecting the larva of the
honeycomb moth, Galleria mellonella, and storing
them in Petri dishes in an incubator, at 21℃, until the
infected host dies and discolouration occurs. This
process usually takes a week after the initial infection.
When extracted, the EPNs are killed and preserved in
Nematode Identification using Artificial Neural Networks
15
DESS solution (Yoder et al., 2006) and then mounted
onto microscopic slides to begin the image capturing
process. We use a light microscope, with an OPTIKA
camera attached, available in the lab to take the
images. The extraction process and image capturing
differ between the infective juveniles (IJ) and the
adults.
3.3.1 Infective Juveniles
As we are dealing with nematodes from the same
feeding group, namely entomopathogenic nematodes,
there is very little difference between each species at
the juvenile stage as they are still developing. There
is a lack of very distinct features between species,
such as genitalia or a pronounced mouth. However,
there is one distinct difference between species at this
life stage, the length of the IJ’s body.
The extraction process of the IJ from the dead
Galleria mellonella uses the White trap method (White,
1927). To use this method, we place the Galleria
mellonella on a small platform, covered in filter paper,
in a container. Water surrounds this platform and soaks
some of the filter paper to create a way for the IJs,
emerging from the cadaver, to enter the water.
To capture images of the IJs, we use a 20x
magnification level on the light microscope. This
magnification level ensures that there is a full focus on
the nematode while also decreasing the amount of
background and any noise visible. There is a total of
188 images in the IJ dataset. The dataset comprises 50
images of Heterorhabditis bacteriophora, 72 images
of Steinernema carpocapsae and 66 images of
Steinernema feltiae.
3.3.2 Adults
The extraction process of the adult nematodes
requires dissecting the dead Galleria mellonella in a
solution of dissolved salts, known as Ringer’s
Solution. This process is necessary as the adult
nematodes never leave the infected host’s body. Once.
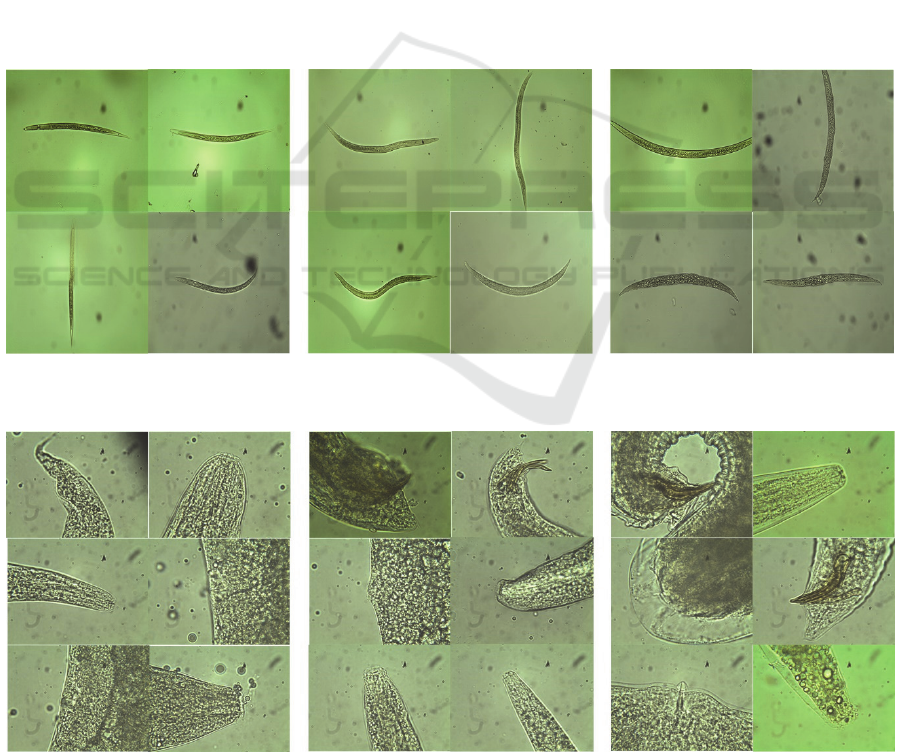
Figure 3: Sample images of infective juveniles from each species at 20x magnification: Heterorhabditis bacteriophora (left),
Steinernema carpocapsae (middle), Steinernema feltiae (right).
Figure 4: Sample images of adult nematodes from each species at 100x magnification:Heterorhabditis bacteriophora (left),
Steinernema carpocapsae (middle), Steinernema feltiae (right).
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
16

dissected, we fish out the nematodes from the cadaver,
using a dissecting needle, and transfer them to a
separate dish of Ringer’s Solution.
To capture images of the adults, we use a 100x
magnification on the light microscope in order to
identify specific features of the nematode. Specific
features of interest are the head, tail and genitalia of the
nematode, the vulva for the female and the spicule for
the male. This dataset contains a total of 234 images.
The images vary in specific features of the nematode
present, as shown in Figure 4. This dataset contains 81
images of Heterorhabditis bacteriophora, 96 images
of Steinernema carpocapsae and 57 images of
Steinernema feltiae.
3.4 Image Pre-processing
3.4.1 Image Size
The acquired images have an original image size of
2048x1536 pixels. This size would require a large
amount of memory and would take a long time to train.
Therefore using the original full size is not feasible.
To make the images more manageable, we reduce the
image to a size of 224x224 pixels as they load in using
the PIL library available for the python programming
language.
3.4.2 Rotations
Considering that both nematode datasets contain a
small number of images, we use methods to generate
more samples for the CNN model. When under a
microscope, nematodes can vary in position and
orientation on the microscope slide. However, CNNs
have issues with rotated objects as the features
learned by a CNN are not rotation invariant. An
example of this issue would be that if a CNN were to
be trained on a set of images and evaluated on the
same set of images flipped upside-down, the CNN
would not be able to make accurate predictions. To
solve this issue, we apply random rotations to the
images as they load in. We also apply random vertical
and horizontal flips to the images with a 50% chance
of applying each flip.
3.4.3 Pixel Scaling
The final step in pre-processing the images is to scale
the pixels down to a smaller range of numbers. This
process reduces large computations and allows for
faster convergence. As we are dealing with image
pixel values, they range from 0 to 255. Using a
method available in the Keras library, for use with the
ImageNet dataset, these values get scaled down to a
range of -1 to 1, achieved by dividing the values by
half of the maximum pixel value, which is 127.5, and
then subtracting by 1.
3.5 Development Environment
To develop and train our CNN models, we employ the
use of a Dell Inspiron 15 7000 Series laptop with an
NVIDIA GTX 960M GPU (4 GB VRAM), an Intel
i7-6700HQ CPU and 16 GB of RAM. This low
amount of VRAM available is taken into
consideration while training, reducing the training
batch size when required to avoid running out of
memory during training.
3.6 Model Training
Using this Xception model, we explore the best
possible method to train on our datasets. To do this
we use three different training methods: Feature
Extraction, Fine-Tuning and Random Initialisation.
Feature Extraction and Fine-Tuning are methods
referred to as Transfer Learning, as they use a pre-
trained model to speed up training and convergence
due to existing features already from a separate
dataset. In the case of the Xception model, the dataset
used is ImageNet (Chollet, 2017). In contrast,
Random Initialisation uses the Xception model with
random weights and bias values, so the model gets
trained from scratch.
The use of those three different training methods
results in training three different models, for both
nematode life stage, and evaluating their performances.
However, we also explore the use of different
optimisers and techniques to find the best combination
for our task. We use SGD, RMSProp and Adam as the
three optimisers for comparison. The other techniques
we apply are Gradient Clipping and Label Smoothing
to see the difference made by applying them versus not
applying them.
Gradient Clipping is an optimiser specific
technique used to avoid large gradient values, also
known as exploding gradients. Label Smoothing is a
technique applied to the one-hot array target output,
decreasing the value of the actual label by a small value
and increasing the values of the other labels by that
small value divided by the number of other labels. We
use a value of 2 for gradient clipping and a value of 0.1
for label smoothing when these techniques are in use.
The effect that label smoothing has on the model is
that it decreases or smoothes out the model’s prediction
distributioin (Pereyra et al., 2017). This effect leads to
the model becoming less overly confident and more
Nematode Identification using Artificial Neural Networks
17
able to generalise. Lastly, we add additional layers to
the model before the output layer to both explore
whether the addition helps or hinders performance and
to provide more trials for better comparisons of other
techniques applied.
3.6.1 Hyper-Parameter Tuning
Each model trained takes roughly one to three hours
to train on our GPU. The variation in training times is
due to the implementation of early stopping to the
models. To allow our models a chance to converge,
we set the total number of epochs to train to a high
value of 2000 epochs. We also set the patience of the
early stopping monitor to 150 epochs, so if the model
does not achieve a lower validation loss than its best
in that time, the training will stop. The best validation
loss for each model is recorded to a spreadsheet for
later comparison. Models that crash during training
are tested again to confirm that the crash occurred due
to memory limitations.
We use a simple bash script to control the training
process, passing in arguments to the python program to
indicate which hyper-parameter settings are to be used.
These hyper-parameters include: the three choices of
training methods, the use of label smoothing, the use of
gradient clipping and the three choices of optimiser.
This resulted in 36 different models being trained for
each nematode life stage, generating a total of 72
models.
The training process was semi-automated as the
addition of model layers required a manual change to
the model and allowed for an assessment of the
progress before training continued. The additional
layers are applied between the global average pooling
layer of the Xception model and the final dense layer.
The additional layers include Dropout, Batch
Normalisation, and Dense layers. The number of units
(neurons) used for the dense layer are based on the
output of the global average pooling layer, which is
2,048 units. Therefore, the models were trialled using
a dense layer with half the number of units, the same
number of units and double the number of units. Other
changes tested included changing the gradient clipping
value and the early stopping patience value.
Overall, each additional change to the model
required training of all 72 models. The total number of
changes to the model explored resulted in 11 different
changes leading to a total of 792 different models
trained. Following the training process, the
misclassification percentage was calculated for each
model using a separate script on a set of non-nematode
images and results were recorded to the spreadsheet.
4 RESULTS
For our results, we measure two different types of
metrics, the validation measures (accuracy and loss)
and the misclassification percentage. To calculate the
misclassification percentage of a model, we get the
model’s predictions on a set of 5000 non-nematode
images. We then use a threshold value of 0.8 on the
softmax outputs and mark the output as incorrect if
any of the label predictions exceed the threshold
value. This measure uses an extreme case to
determine whether a model is too confident on the
images of nematodes that it will give a high prediction
of a nematode label even if a nematode is not present
in the image.
Some models would crash during their training due
to the memory required exceeding the total amount of
memory available. We omit these models from our
analysis. More often, models that were prone to
crashing were the models that had additional layers
added. This fact is more due to the memory limitations
of our hardware. However, a surprising effect shown
was that models trained using Adam as the optimiser
crashed significantly more often than models using the
other optimisers.
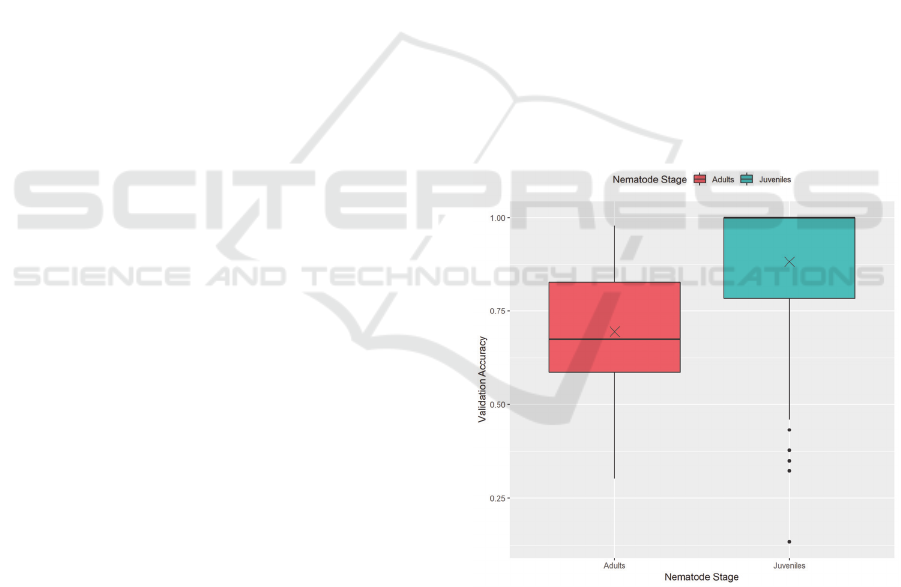
Figure 5: Validation accuracy by nematode stage.
Figure 5 shows the difference in validation
accuracy performance between the juvenile dataset and
the adult dataset. This figure includes all recorded
training results, including all training methods. This
figure shows that models trained on the IJ dataset
achieved significantly higher accuracies than ones
trained on the adult dataset. The cause for this is most
likely due to the difference in features of interest
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
18
between the two. The X marks the mean validation
accuracies for each nematode stage.
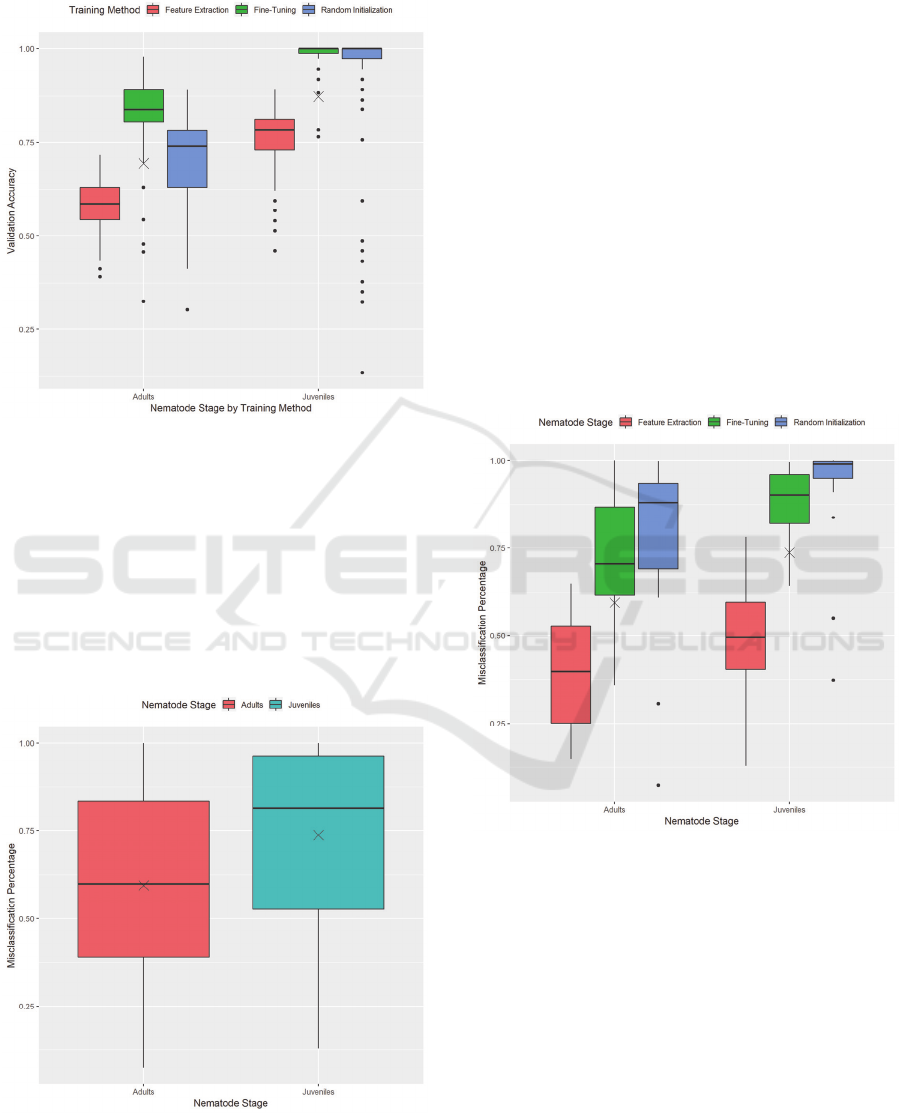
Figure 6: Validation accuracy by nematode stage, separated
by training method.
Separating these results by training method
provides us with Figure 6. This figure gives us a more
precise image as to how each training method affects
the performance of a model. With both nematode
stages, feature extraction leads to the lowest
performance accuracy, while fine-tuning leads to the
highest performance accuracy, often staying at 100%
for the IJ dataset.
Figure 7: Misclassification Percentage by Nematode Stage,
excluding results using Label Smoothing and Gradient
Clipping.
The adult dataset presents a vast range of values
for validation accuracy across all training methods.
This fact is most likely due to the small amount of
variation in the adult nematode dataset. Rotations do
provide enough images to train on; however, more
samples of nematodes would help increase the
variation and lead to less varied performance
between models.
As the models present the ability to perform
adequately on both datasets, we now look at how these
models perform in an extreme case using non-
nematode images. Figure 7 shows that both datasets
have an average misclassification percentage of over
50%, meaning that most models incorrectly predicted
more than half of the non-nematode images as a
nematode. When explored carefully, the models appear
to have a default label they predict, which presents a
possibility of over-fitting. We will now show how each
training method affects this misclassification
percentage.
Figure 8: Misclassification percentage by training method,
excluding results using Label Smoothing and Gradient
Clipping.
Figure 8 presents a more precise image and
allows us to understand what is happening. While
feature extraction showed to have a generally lower
accuracy compared to the other training methods, it
shows here to have a lower level of misclassification
on non-nematode images. This fact is due to the
preservation of the weights of all the layers from the
pre-trained Xception model while training. The
learned ImageNet features allow for the model to be
less likely to label a non-nematode image as a
nematode.
Nematode Identification using Artificial Neural Networks
19
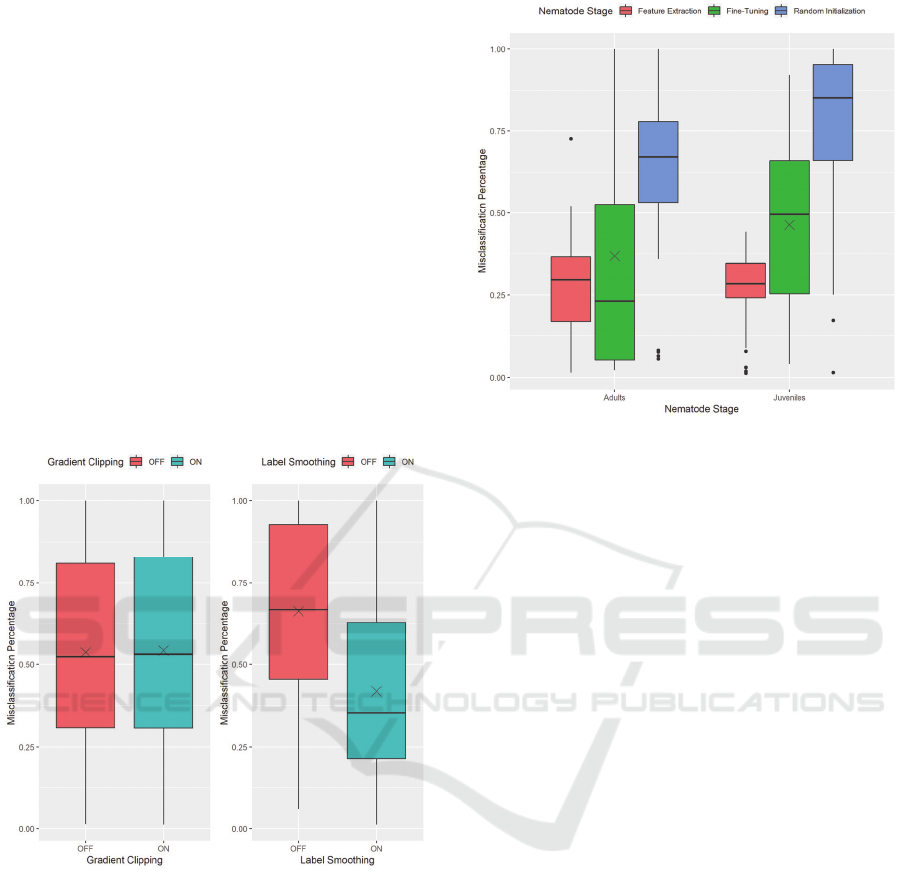
Random initialisation performs the worst, evident
by the IJ dataset having very high misclassification
percentages, due to these models training from scratch
with no pre-existing knowledge to use. However, Fine-
Tuning shows high misclassification percentages as
well, even with pre-existing knowledge. Our
implementation of fine-tuning allows all layers to have
their weights updated during training. This
implementation alters the pre-existing feature
extraction leading to a significant variation in
misclassification percentage values.
To combat this issue, we investigated using some
techniques applied in some state-of-the-art models:
Label Smoothing and Gradient Clipping (Szegedy et
al., 2016; Zoph et al., 2017). Upon first discovering the
trend of misclassification, we selected these methods
to test if models were exhibiting exploding gradients
and if models were becoming overly confident with
their predictions.
Figure 9: The effects of Gradient Clipping (left) and Label
Smoothing (right) on misclassification of non-nematode
images.
We can see the effects that these techniques have
on the misclassification percentage in Figure 9. On the
left, it shows that gradient clipping does not affect the
level of misclassification exhibited by the models.
However, label smoothing shows a considerable
decrease in the average misclassification of a model on
non-nematode images.
Figure 10: Misclassification percentage by training method,
using Label Smoothing.
To further show the effect of label smoothing, we
have Figure 10 presenting the misclassification
percentage of models using label smoothing. This
figure is comparable to that of Figure 7. Figure 10
presents a considerable decrease with all training
methods. This decrease is especially evident with Fine-
Tuning and Random Initialisation, both having had
very high misclassification percentages without the use
of label smoothing.
4.1 Comparisons
As there is no standard benchmark for the task of
identification, it is difficult to compare with other
approaches that use similar techniques. Other
researchers have used similar techniques, but for
different purposes such as detection, counting and
identification based on behavioural dynamics rather
than morphological features. However, we see a
significant improvement compared to cases where
deep learning has been used for tasks involving
nematodes including identification, detection and
counting.
Other image classification models have been
trained with a more substantial number and variety of
images, such as the Xception model which achieved a
79% Top-1 accuracy on the ImageNet dataset (Chollet,
2017). Although we make use of the Xception model
architecture, in comparison, our dataset is small in
quantity and variation. Despite this, it still performs
well. Our models achieved an average validation
accuracy of 88.28% for the juveniles dataset and
69.45% for the adult dataset.
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
20

5 CONCLUSION
From our analysis, we show that while there is no
single best model for performing our specific task,
there are many techniques that show improvements to
performance over others. The use of Fine-Tuning
provides our model with existing knowledge, from
the ImageNet dataset, to speed up training and allow
for faster convergence. However, these models risk
becoming overly confident and misclassifying non-
nematode images at a high rate. With the use of Label
Smoothing, these models are less likely to make
incorrect predictions on non-nematode images, as
they become more able to generalise.
As this study was dealing with nematodes from the
same trophic group and even two nematode species
from the same family, this technology has shown no
issues in being able to differentiate between them. This
is an achievement for the technology, as often there are
only minuscule differences between species, especially
species belonging to the same family. While we used
nematodes from the same trophic group, an
investigation will be required into how well this
technology will scale to more nematode species, due to
thousands of different species of nematodes existing
and with millions more estimated to exist.
There is a need for more images, both of the ones
used in this study and of other nematode species, to
determine how well this technology will scale.
Utilising images from other nematode researchers
would provide a variety of types of nematodes and a
variation in images. This can also cut down on any time
required for data gathering, as culturing nematodes is
very time-consuming. Creating a sizeable standard
dataset with these images would also provide more
opportunities to explore improving Nematode
Identification with Deep Learning and any other
advancing technology.
As most approaches using computers to aid in
Nematode Identification have failed to be adopted by
an audience other than the authors themselves, we hope
that increased research could help improve the state of
computer-aided approaches. These approaches not
only being Nematode Identification but many other
tools, such as counting, to improve the analysis of these
organisms.
REFERENCES
Akintayo, A., Lee, N., Chawla, V., Mullaney, M., Marett,
C., Singh, A., Singh, A., Tylka, G.,
Ganapathysubramaniam, B., & Sarkar, S. (2016). An
end-to-end convolutional selective autoencoder
approach to Soybean Cyst Nematode eggs detection.
arXiv preprint arXiv:1603.07834.
Akintayo, A., Tylka, G. L., Singh, A. K.,
Ganapathysubramanian, B., Singh, A., & Sarkar, S.
(2018). A deep learning framework to discern and count
microscopic nematode eggs. Scientific reports, 8(1), 1-
11.
Bouket, A. C. (2012). NEMIDSOFT: A Web-based
Software as an Aid to the Identification of Nematodes.
International Journal of Computer Applications, 57(4).
Chollet, F. (2017). Xception: Deep learning with depthwise
separable convolutions. In Proceedings of the IEEE
conference on computer vision and pattern recognition
(pp. 1251-1258).
Cortet, J., Gomot-De Vauflery, A., Poinsot-Balaguer, N.,
Gomot, L., Texier, C., & Cluzeau, D. (1999). The use
of invertebrate soil fauna in monitoring pollutant effects.
European Journal of Soil Biology, 35(3), 115-134.
Culurciello, E. (2018, December 24). Neural Network
Architectures. Medium. https://towardsdatascience.
com/neural-network-architectures-156e5bad51ba
Dargan, S., Kumar, M., Ayyagari, M. R., & Kumar, G.
(2019). A Survey of Deep Learning and Its
Applications: A New Paradigm to Machine Learning.
Archives of Computational Methods in Engineering, 1-
22.
de Oliveira, R. C., do Nascimento Queiroz, S. C., da Luz,
C. F. P., Porto, R. S., & Rath, S. (2016). Bee pollen as
a bioindicator of environmental pesticide
contamination. Chemosphere, 163, 525-534.
Diederich, J., Fortuner, R., & Milton, J. (2000). Genisys and
computer-assisted identification of nematodes.
Nematology, 2, 17–30.
Dillman, A. R., & Sternberg, P. W. (2012).
Entomopathogenic nematodes. Current Biology, 22(11),
R430-R431.
Dodds, W. K., & Whiles, M. R. (2010). Chapter 10—
Multicellular Animals. In W. K. Dodds & M. R. Whiles
(Eds.), Freshwater Ecology (Second Edition) (Second
Edition, pp. 221–257). Academic Press.
Fortuner, R. (Ed.). (2013). Nematode identification and
expert system technology (Vol. 7). Springer Science &
Business Media.
Goodfellow, I., Bengio, Y., & Courville, A. (2016). Deep
learning. MIT press.
Javer, A., Brown, A. E., Kokkinos, I., & Rittscher, J. (2018).
Identification of C. elegans strains using a fully
convolutional neural network on behavioural dynamics.
In Proceedings of the European Conference on
Computer Vision (ECCV) (pp. 455-464).
Kalwa, U., Legner, C., Wlezien, E., Tylka, G., & Pandey,
S. (2019). New methods of removing debris and high-
throughput counting of cyst nematode eggs extracted
from field soil. PloS one, 14(10).
Kennedy, A. C., & Luna, L. Z. de. (2005). RHIZOSPHERE.
In D. Hillel (Ed.), Encyclopedia of Soils in the
Environment (pp. 399–406). Elsevier.
LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998).
Gradient-based learning applied to document
Nematode Identification using Artificial Neural Networks
21

recognition. Proceedings of the IEEE, 86(11), 2278-
2324.
LeCun, Y., Bengio, Y., & Hinton, G. (2015). Deep learning.
nature, 521(7553), 436-444.
Milton, J., Fortuner, R., & Diederich, J. (2000). Genisys and
computer-assisted identification of nematodes.
Nematology, 2(1), 17-30.
Poinar, G. O. (2016). Chapter 9—Phylum Nemata. In J. H.
Thorp & D. C. Rogers (Eds.), Thorp and Covich’s
Freshwater Invertebrates (Fourth Edition) (Fourth
Edition, pp. 169–180). Academic Press.
Pereyra, G., Tucker, G., Chorowski, J., Kaiser, Ł., & Hinton,
G. (2017). Regularizing neural networks by penalizing
confident output distributions. arXiv preprint
arXiv:1701.06548.
Seesao, Y., Gay, M., Merlin, S., Viscogliosi, E., Aliouat-
Denis, C. M., & Audebert, C. (2017). A review of
methods for nematode identification. Journal of
microbiological methods, 138, 37-49.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna,
Z. (2016). Rethinking the inception architecture for
computer vision. In Proceedings of the IEEE
conference on computer vision and pattern recognition
(pp. 2818-2826).
Tan, M., & Le, Q. V. (2019). Efficientnet: Rethinking
model scaling for convolutional neural networks. arXiv
preprint arXiv:1905.11946.
Tsang, S.-H. (2019, March 20). Review: Xception — With
Depthwise Separable Convolution, Better Than
Inception-v3 (Image Classification). Medium.
https://towardsdatascience.com/review-xception-with-
depthwise-separable-convolution-better-than-
inception-v3-image-dc967dd42568
White, G. F. (1927). A method for obtaining infective
nematode larvae from cultures. Science (Washington),
66(1709).
Wilson, M. J., & Khakouli-Duarte, T. (Eds.). (2009).
Nematodes as environmental indicators. CABI.
Xie, Q., Hovy, E., Luong, M. T., & Le, Q. V. (2019). Self-
training with Noisy Student improves ImageNet
classification. arXiv preprint arXiv:1911.04252.
Yoder, M., King, I. W., De Ley, I. T., Mann, J., Mundo-
Ocampo, M., Poiras, L., De Ley, P., & Blaxter, M.
(2006). DESS: a versatile solution for preserving
morphology and extractable DNA of nematodes.
Nematology, 8(3), 367-376.
Zoph, B., Vasudevan, V., Shlens, J., & Le, Q. V. (2018).
Learning transferable architectures for scalable image
recognition. In Proceedings of the IEEE conference on
computer vision and pattern recognition (pp. 8697-
8710).
DeLTA 2020 - 1st International Conference on Deep Learning Theory and Applications
22
