
Holistic Health Records towards Personalized Healthcare
Athanasios Kiourtis
1
, Argyro Mavrogiorgou
1
, Dimosthenis Kyriazis
1
, Francesco Torelli
2
,
Domenico Martino
2
and Antonio De Nigro
2
1
Department of Digital Systems, University of Piraeus, Piraeus, Greece
2
Engineering Ingegneria Informatica, SpA, R&D Laboratory, Rome, Italy
Keywords: CrowdHEALTH, HL7 FHIR, Holistic Health Records, Interoperability, Personalized Healthcare.
Abstract: Current healthcare systems include platforms that provide data not linked to each other. However, linking
clinical information to other people’s life data would be beneficial in understanding the effects of prevention
strategies, and diseases. More specifically, in a context with several data sources, setting of a baseline allowing
the aggregation of information, avoiding ambiguities, is crucial. This manuscript presents the Holistic Health
Records (HHRs), as health records that intend to provide a complete picture of a patient, including all health
determinants. This data may be produced by different systems at different times of the patient’s life, including
data related to regular patientcare and non-medical data that may affect the patient’s state of health. Many
standards have been defined with this purpose, with HL7 Fast Healthcare Interoperability Resources (FHIR)
being mostly tailored to the current needs. Hence, the HHR model based on HL7 FHIR has been designed,
representing information about persons, their roles, their healthcare organizations, diagnosis and clinical
findings of the patients, among others. The HHR model aims on guaranteeing interoperability and being
implemented on top of existing FHIR libraries and is also intended to be usable independently from FHIR
and applicable for different purposes than only exchanging health data.
1 INTRODUCTION
The explosion of healthcare services is leading to the
creation of several devices and platforms that, one
independently from the others, provide data on the
citizen’s life (Mavrogiorgou, 2017). Linking clinical
information with other citizens’ life data would be of
benefit for learning about outcomes of prevention
strategies, diseases, and efficiency of clinical
pathways (Kiourtis, 2019). The challenge is to
combine all the available data to exploit the
community knowledge benefits, by building the
“Holistic Health Records” (HHRs) that include any
information that is relevant to a citizen’s health (i.e.
medical, clinical, lifestyle, social care data, etc.).
Even though thousands of medical data models exist
(Geßner, 2017), they are mainly focused on the
integration of data from clinical trials. More general
interoperability data models (e.g. OpenEHR
(openEHR, 2021), HL7 Fast Healthcare
Interoperability Resources (HL7 FHIR) (HL7 FHIR,
2021) are sufficiently general and extensible to cover
holistic needs, but do not guarantee a full
homogeneity, as they leave freedom to represent the
same information using different data structures or
different coding systems.
The data model of the HL7 FHIR revolves around
a series of interoperability artefacts composed of a set
of modular components called “resources”. These
resources are discrete information units with defined
behaviour and meaning, describing what information
can be collected for each type of clinical information.
Currently, there exist different resources for
structuring information from a patient, an adverse
reaction, a procedure, and an observation, among
many others. Within FHIR, the different types of
resources can be classified in six major categories: (a)
Clinical: content of clinical record, (b) Identification:
supporting entities involved in the care process, (c)
Workflow: management of the healthcare process, (d)
Financial: resources that support the billing and
payment parts of FHIR, (e) Conformance: resources
that manage specification, development and testing of
FHIR solutions, and (f) Infrastructure: general
functionality and resources for internal FHIR
requirements. The content of the FHIR resources can
be represented in different formats such as XML,
JSON and Turtle, although other formats are also
78
Kiourtis, A., Mavrogiorgou, A., Kyriazis, D., Torelli, F., Martino, D. and De Nigro, A.
Holistic Health Records towards Personalized Healthcare.
DOI: 10.5220/0010509300780089
In Proceedings of the 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2021), pages 78-89
ISBN: 978-989-758-506-7
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

allowed. Consequently, it is possible to obtain
information structured according to the FHIR
resource data model, and represented in one of these
formats, resulting that this information can be
readable by both humans and machines. Within this
standard, 119 other resources (apart from the patient
resource) are defined at different maturity levels. To
this context, HL7 aims to define and limit the
structures used for the exchange of clinical
information.
Regarding the different coding systems, while a
terminology can refer to several different things, in
healthcare it is associated with the “language” used to
code entries in Electronic Health Records (EHRs)
(Monsen, 2019) including LOINC (LOINC, 2021),
SNOMED-CT (SNOMED-CT, 2021), ICD-10 (ICD-
10, 2021), or ICD-9 (ICD-9, 2021), among others.
Most people encounter medical terminologies at
some point in their lives – whether it is as physicians,
medical purchasers, or patients. In the world of EHRs,
terminology is one of the key parts for achieving real
interoperability between healthcare systems and
integrating their data. For instance, in the case that it
is needed to send data between two systems, for the
data to be usable, these systems must “communicate”
in the same language. This means that the codes from
one system must be compatible with the codes from
the other system. While it can be easy to combine data
from multiple systems in one place, in the case that
these codes cannot be mapped to one another, then the
data remain locked (Mavrogiorgou, 2017). Currently,
there exist several standards. As a result, a lot of
research is performed to map these various
vocabularies so that one can move easily from one to
the other, as long as one of the key ones listed earlier
is used. To this end, there is work that has been done
and is ongoing, such as mappings between ICD-9 and
ICD-10, LOINC and CPT (CPT, 2021), or LOINC
and SNOMED CT.
In this context, it should be noted that medical
information is typically represented following some
specific standards. The SNOMED-CT terminology is
an ontology that defines (some) concepts, such as,
some diseases in terms of their cause, the part of the
body they affect and how they can be diagnosed. It
also includes some food categories, sport categories
or activities of daily living. The Open Biomedical
Ontologies (OBO) consortium (Smith, 2007) is an
initiative trying to integrate the multiple ontologies
developed in the biomedical domain, which also
includes ontologies formalizing patient medical care
and EHRs. The International Classification of
Functioning, Disability and Health (ICF) (Cieza,
2002) is an ontology classifying health and health-
related domains from a body perspective, a personal
activities perspective and a societal perspective. It
classifies according to the body structure (i.e. eye,
ear, digestive systems, etc.), the body function (i.e.
mental, voice, etc.), activities and participations and
the environmental context. Thus, it contains medical
categories as well as some social categories as part of
the activities, participations, and environmental
domains. All concepts are linked to the ICD code in
the ICD terminology. The National Cancer Institute
Thesaurus (NCIT) (Zhe, 2002) is a reference
thesaurus covering biomedical concepts and inter-
concept relationships. As part of that, it also includes
medical categories, categories for physical activities,
social activities and behavioural categories.
However, a major problem is the success of using
ontologies in many domains, as it leads to the
development of many different not necessarily linked
ontologies and taxonomies. This creates in practice
the problem of interoperability, both at the taxonomic
and the semantic levels. To overcome that problem,
major effort is provided from initiatives, such as OBO
and BioPortal (Noy, 2009). It is also the motivation
for the OntoHub (Mossakowski, 2014) repository,
which behind the scenes attempts to utilize alignment
techniques from formal methods for the ontology
domain. The Medical Subject Headings (MeSH)
(Lipscomb, 2000) is a vocabulary maintained by the
US National Library of Medicine (NLM) (Lindberg,
1990). It is a hierarchically organized terminology of
biomedical information contained in NLM database,
including MEDLINE®/PubMed® (Fontelo, 2005). It
is often combined information following the RxNorm
(Liu, 2005), as well as the LOINC standard for
medical laboratory observations. Therefore, the mere
adoption of interoperability standards is not sufficient
to query health data coming from various health data
sources and systems, in a uniform, efficient,
complete, and unambiguous way.
In this manuscript, it is presented the data
integration approach in the form of HHRs that has
been adopted by CrowdHEALTH (Kyriazis, 2017).
CrowdHEALTH is a digital healthcare system aiming
to exploit big data techniques, applied to extended
health records and collective health knowledge (i.e.
clustered records), to evaluate healthcare governance
policies. One of the pillars of the CrowdHEALTH
system is the development and exploitation of the
HHRs. HHRs are intended to provide an integrated
view of the patient, including all health determinants.
Such health-related data may be produced by
different human actors or systems, in different
moments of a patient’s life, and include both i)
medical data, associated with regular patientcare or a
Holistic Health Records towards Personalized Healthcare
79

part of a clinical program, and ii) non-medical data
that may have an impact on the patient’s health status.
HHRs potentially include: (a) social and lifestyle data
collected by either the patient or other individuals
(e.g. family members, friends), (b) social care data
collected from social care providers, (c) physiological
and environmental data collected by medical devices
and sensors, (d) clinical data coming from healthcare
information systems and produced by healthcare
professionals (e.g. primary care systems and
electronic medical records), (e) laboratory medical
data, and (f) nutrition data.
The rest of this document is structured as follows.
Section 2 describes the HHR model, its goal and the
approach followed to realize it, aiming at satisfying
various data requirements, while Section 3 reports the
experimentation, the evaluation and the development
steps followed towards the creation of the HHR
model, through an easily followed example. To this
end, Section 4 includes an overall discussion of the
current results, including our conclusions and next
steps.
2 METHOD
2.1 Main Principles of the HRR Model
The goal of the CrowdHEALTH personalized
healthcare system is the development of a set of data
analysis tools that can be applied to different use
cases, possibly merging data coming from different
contexts. Therefore, there is the need to define one
integrated model for HHRs, in order to guarantee the
possibility to apply these tools to all produced data.
For these reasons, firstly the HHR model has to
represent in a consistent way all the data required by
the specific use cases. Secondly, the model is
intended to be a seed for future extensions. Thus, it
includes also types of data that are not currently
required but are considered likely to be used in the
near future or are useful to exemplify how the model
can be extended in the future. Thirdly, the model is
defined using existing models as a reference. In
particular, the FHIR standard has been selected as the
main reference for the definition of the HHR model.
While this standard is still under development and is
mainly capable to represent clinical data, it already
includes the possibility to represent data that is not
necessarily clinical, such as information coming from
environment sensors or related to the social aspects.
Moreover, thanks to the adoption of the concept of
“resources” and the definition of a flexible extension
mechanism, the FHIR model is conceived from the
fundament to be applicable in different contexts.
Together with the FHIR standard, CrowdHEALTH
also considers ontologies at the state of the art, useful
to qualify entity types that correspond to
specializations or abstractions of entities represented
by FHIR elements. Fourthly, the HHR model is
designed at conceptual level and in parallel mapped
with existing standards. The model is provided both
in a semiformal format, using UML, and in a
completely formal format, using a tiny XML
language, the HHR mapping language that was
created in the context of CrowdHEALTH. The HHR
mapping language allows to express in a simple way
both the structure of the model and its mapping to
FHIR and to existing or new terminologies. Several
constraints are imposed to the designer of the HHR
model to guarantee the feasibility of a direct mapping
to FHIR. The reason for not using directly the
selected reference standard is to untie the HHR model
from choices related only to the FHIR
implementation (for instance to simplify the
implementation of restful services), and make explicit
in the model some aspects that are implicit in FHIR,
so to ease the usage of the HHR model independently
from FHIR. Therefore, the HHR model aims on the
one hand to be easily implementable on top of
existing FHIR implementations, and on the other
hand it is also intended to be easily implementable
using different technologies. For example, the
CrowdHEALTH systems uses a Java implementation
of the HHR model, which is automatically generated
using the HHR mapping language and is different
from FHIR implementations, although easily
convertible to it.
2.2 Organization of the HHR Model
Similarly, to some of the existing standards, the HHR
model is described in a semi-formal way using UML.
Differently from other models, the HHR model also
has a completely formal description expressed with
the HHR mapping language (not described in this
manuscript). The overall model is divided in several
packages to simplify the representation and the
description of the reported information. Each package
collects information related to a specific topic (e.g.
the representation of the information characterizing a
Person, clinical Conditions of patients or
Measurements performed on Persons). For each
fragment, the description of each entity and its
relationships with the other entities in the fragment is
reported. Although the model is split in several
packages, its classes belonging to different packages
have always different names, in order to reduce the
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
80

risk of misunderstanding and enable also
implementations that put all classes in a single
software package.
2.3 Level of Abstraction and Scope of
the HHR Model
As a rule, each class of the HHR model corresponds
to a resource type or a data type of the FHIR model,
with the difference that the HHR model is designed at
an ontological level and is more specialized than the
FHIR model. The usage of an ontological approach is
in particular evident in two aspects that distinguish
the HHR model from the FHIR model. One aspect is
that the multiplicity constraints on the UML
associations and attributes do not represent integrity
constraints, as in the case of FHIR, but represent real
world existence constraints. For instance, if an
attribute has minimum multiplicity equal to 1, this
does not imply that the value of that attribute must be
mandatorily stored or transmitted when exchanging
data, but only implies that at least one value of that
attribute always exists in the world, and this
information is actually not stored in any information
system or not transmitted. All the attributes of the
entities in the HHR model are not mandatory, i.e.
their values are not required to be stored or
transmitted for each data transmission occurrence.
Another aspect is the usage of abstract classes that
have no direct corresponding type in FHIR but
correspond to super-types of FHIR resource types.
Such classes are introduced to make explicit some
semantic commonalities that are implicit in the FHIR
model. Moreover, in order to represent ontological
distinctions that cannot be expressed with standard
UML, a specific stereotype and pattern is adopted.
For example, classes of entities (e.g. Patient) that
correspond to roles of instances of other classes (e.g.
Person), are marked with the stereotype <role> and
use the standard relation “player” to associate the
entity (e.g. the person) that plays the role. If needed,
implementations of the HHR model may exploit the
explicit representation of roles and accept to assign
instances of a certain role as a value of attributes
whose type is not that role, but it is the type of the
instances that may play that role (e.g. accepting a
Practitioner as value of an attribute expecting a
Person). However, this cannot be realized for the vice
versa scenario (i.e. it is forbidden to assign a Person
to an attribute expecting a Practitioner). When a class
C has numerous subclasses, but these subclasses add
no specific attributes or constraints, then the
subclasses are reified. Each subclass is represented by
an item of an enumeration (stereotype <enum>) and a
mandatory attribute of the class C (with name Ctype)
is used to represent the specific subclass of the
instance. For example, the subclasses of the class
Condition correspond to values of the enumeration
ConditionType and the specific subclass of a
Condition instance is represented by the value of the
attribute named conditionType. The fact that the
HHR model is more specialized than the FHIR model
is also evident in several aspects. The most important
aspect in the HHR model is the absence of classes and
elements that are present in FHIR, since they are not
needed by the current CrowdHEALTH use cases.
Moreover, an HHR class that corresponds to a certain
FHIR resource class may have explicit subclasses that
are not represented as distinct resource classes in
FHIR. Differently from the addition of new attributes,
usually the introduction in the HHR model of these
explicit subclasses does not require a corresponding
FHIR extension. The instances of all such HHR
subclasses correspond to instances of the same FHIR
resource class, and their semantic type is distinct by
assigning a specific value, chosen from specific
terminologies, to a “category” or “code” attribute of
the resource class. The values of these attributes are
fixed by the HHR model, in order to assure that the
same type of data is always represented using the
same terms from the same terminologies. In other
terms, the HHR model explicitly and unambiguously
represents concepts that are needed by the
CrowdHEALTH use cases and either are implicit in
FHIR or need a FHIR extension.
As mentioned above, a few constraints are
imposed to the HHR model to guarantee an easy
mapping with FHIR and with specific coding
systems. The main constraint is that any leaf element
of the HHR model (i.e. any class, attribute or
association that does not have subclasses or
specializations) must correspond to exactly one
(resource or data) type of the FHIR model, i.e. all
possible instances of an HHR class must represent the
same entities of possible instances of only one
corresponding FHIR class. Another constraint is that
each instance of an HHR class must correspond to
exactly one instance of the FHIR model. On the other
hand, any non-leaf element of the HHR model, is
considered ontologically “abstract” (i.e. all its
representable instances or values must be instances or
values of some subclass). This is intended to avoid the
usage of instances of non-leaf classes to represent
unintended entities. Implementations may impose the
instantiation of only leaf classes. As HHR classes are
conceptual, advanced implementations may also
allow to instantiate non-leaf classes of the HHR
model, in order to allow to represent entities whose
Holistic Health Records towards Personalized Healthcare
81

type is not completely known, possibly allowing to
specify a more specific type in a second moment (i.e.
allowing the same instance to conceptually move
from a superclass to a subclass when more
information is available). Although the semantics of
the HHR elements are usually more specific than the
ones of the FHIR model, in order to make the
mapping more evident, the name of the most general
HHR element that is mapped to a specific FHIR
element usually takes the same name of the
corresponding FHIR element. In any case, different
names are chosen when the semantics of the HHR
element are specific and would be misleading to
adopt the same name with FHIR. The detailed
specialization of the HHR model, with respect to
more general-purpose standards, has the advantage of
reducing the ambiguity of the model and simplifying
its comprehension, thus mitigating the risk that
different standard elements are used to represent the
same type of information. The final version of the
HHR model aims to represent the information
enabling the execution of all the use cases of the
CrowdHEALTH system.
2.4 Steps Followed to Define the HHR
Model
Following the general incremental development
approach of the CrowdHEALTH project, the
development of the HHR model followed a multi-
cycles process, producing two different versions of
the HHR model aligned with corresponding versions
of the use case requirements. In each development
cycle, different tasks have been performed.
(i) Firstly, each use case leader has been asked to
describe the information that she would like to store
and analyse using the CrowdHEALTH tools,
focusing on the data needed for the first version of
their use case implementation. A template was
provided to each use case to perform this description.
It was asked to create and describe a UML conceptual
diagram representing the type of entities and
relationships described by their data source
(abstracting from implementation details of the actual
database scheme). It was also asked to describe, using
specific tables, each attribute of each entity and the
corresponding cardinality and value constraints. In
the second cycle this description has been in some
case produced by extending the one produced during
the first cycle, and in some other case, starting the
process from scratch to obtain a better model.
(ii) Secondly, different analysts have been
assigned to each use case, in order to clarify
ambiguity issues related to their data source and to
express a mapping of their dataset scheme to the
FHIR model, in order to disambiguate the semantics
of each type, relationship and attribute. The mapping
was expressed using specific tables and the FHIRPath
language.
(iii) Thirdly, all the conceptual models produced
by the use cases have been merged, one by one, in a
unique HHR model. In this phase, different
conceptual classes that different use cases had
mapped to the same FHIR classes or to FHIR classes
with similar semantics have been merged in a unique
HHR class, or in different subclasses of a same
abstract HHR class. The same analyses have been
performed for attributes and associations.
(iv) Fourthly, it took place the formalization of the
mapping to FHIR using the same semi-formal
approach used for the mapping of data source
conceptual schemes.
(v) Fifthly, it took place the definition of the HHR
model and of the mapping to FHIR using the formal
XML language specified by the CrowdHEALTH
project.
The resulting specification distinguish general
purpose concepts that are included in the HHR model,
and extensions to the HHR model required by specific
use cases. The extensions of the HHR model are
formalized in the same way of the HHR model, but
are not considered mandatory parts of the HHR
model, because they represent information that is
meaningful only to a specific organization and does
not need to be exchanged in a standardized way with
other organizations.
2.5 Health-related Aspects Covered by
the HHR Model
The data types that are covered by the HHR model
belong to nine different categories. In particular:
(i) Physical activities: workouts, biodata and
fitness tests performed by a person or groups of
persons.
(ii) Lifestyle: data concerning sleep, substances
consumption such as alcohol, tobacco or recreational
drugs. It also covers data regarding daily habits.
(iii) Social: data related to social interactions, such
as the emotion, the number of the contact in the phone
or the number of exchanged multimedia items.
(iv) Events: all aspects concerning episode of care,
hospitalizations, clinical procedures, laboratory tests
and care plans.
(v) Medications: all data regarding the
prescription, request, and assumption of medication.
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
82
(vi) Conditions: symptoms, diagnosis, allergies,
and intolerances that a specific patient or group of
patients suffer.
(vii) Nutrition: all data regarding the food and
beverage intake.
(viii) Administrative: demographics and other
administrative information about an individual or
group of individuals. It also includes information
about the educational level, occupational status and
assurance of individuals, and information about
organizations.
(ix) Measurements: measurements and simple
assertions about a patient, device, or other subject. It
also includes collective health measurements about a
group of persons sharing common characteristics.
2.6 Health-related Aspects Covered by
the HHR Model
This sub-section presents the “Person” package of the
HHR model, which specifies classes, attributes and
roles characterizing a person or a group of people.
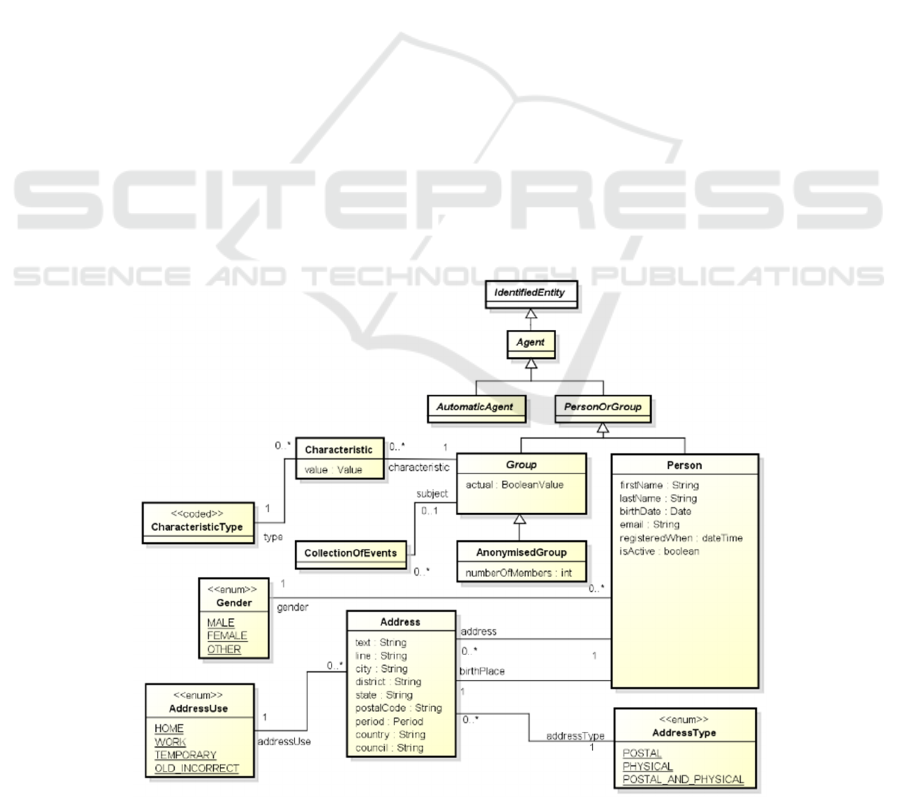
Fig. 1 illustrates a UML class diagram that includes
the class Person of the HHR model, which represents
demographics and administrative information about a
person that is independent of any specific health
context. The gender is modelled by the Gender
enumeration, while her address and birthplace are
modelled with the class Address, having a specific
AddressUse and AddressType. The same figure
illustrates the class Group, which represents a group
of people sharing a common set of characteristics
and/or a common set of CollectionOfEvents. The
reification of a valorised property/attribute is
represented by the class Characteristic, while the
CharacteristicType is the attribute/property that is
reified by a characteristic. Group is specialized in
AnonymisedGroup when the identity of their
members is unknown. Person and Group inherit from
PersonOrGroup together with AutomaticAgent, the
unique identifier from their superclass Agent, from
which a specific person or group of persons may be
identified in CrowdHEALTH.
The Class Coverage (shown in Fig. 2), associated
to Person, is intended to provide the high-level
identifiers and potentially descriptions of an
insurance plan, which may be used to pay for, in part
or in whole, the provision of healthcare products and
services. Its status is modelled with the
FinancialResouceStatus enumeration. A same person
can play different individual roles into different
contexts. Each individual role of the same person is
represented by a different instance of the class
PersonInTime. An instance of PersonInTime is a
view of a person related to a specific time frame
and/or a specific context (depending from the specific
subclass). This means that a same person may
correspond to several instances of PersonInTime,
where each instance describes information of the
person that is specific to the corresponding role, and
Figure 1: HHR demographic attributes characterizing a person.
Holistic Health Records towards Personalized Healthcare
83
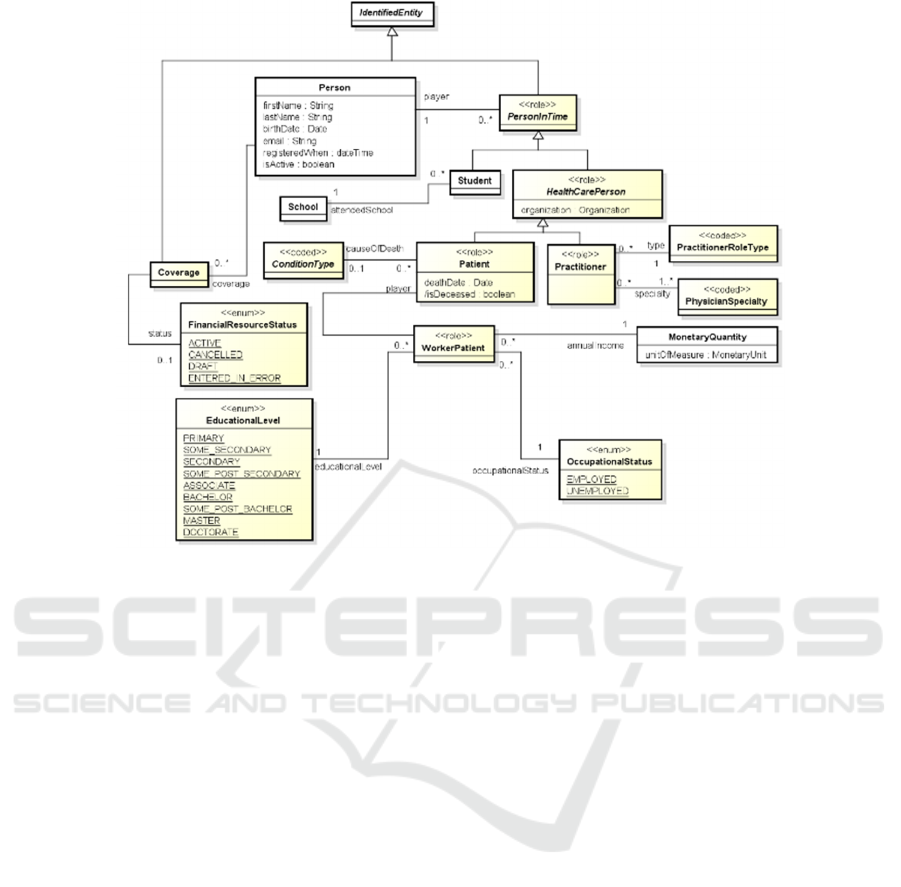
Figure 2: HHR attributes characterizing the roles of a person.
it is related using the “player” association-end, to the
person that plays that role. In particular, a person has
the role Patient when she is the subject of the
healthcare activities provided by HealthCare
professionals.
If the same person has been assisted by two
different healthcare providers, then it plays two
different Patient roles (corresponding to two different
instances of the class Patient). On the other side, a
person has the role of Practitioner when she is a
qualified medical doctor, with one or more
PractitionerSpecialty, and works for a specific
organization. If the same practitioner works for
different organizations, it plays two different
instances of PractitionerRoleType, corresponding to
the set of the roles that a practitioner may perform at
an organization for a specific period. The superclass
of Patient and Practitioner is HealthCarePerson,
representing any person that plays a role in an
HealthCareOrganization. The role of WorkerPatient
is played by a Person that performs a specific work in
a specific frame of time that has an EducationLevel
and an occupationalStatus with a specific annual
income represented by the class MonetaryQuantity.
When a Patient passed away, the cause of the death is
represented by an instance of the class
ConditionType.
3 RESULTS
3.1 Source Code
A specific Java library, called HHR Manager, allows
to instantiate and modify in-memory Java objects that
are compliant to the HHR conceptual model. Based
on what has been described in Section 2, in order to
produce the HHR conceptual model, the HHR model
has been first formalized using a language called
“HHR mapping language”. This is an XML language,
specifically designed for the HHR model, that allows
to specify in a machine-interpretable way the
structure of HHR types and map them to the structure
of corresponding FHIR resources. The HHR mapping
language is basically a declarative language for
defining and mapping document oriented (i.e. tree-
like) data structures and exploits the FHIRPath
language to navigate such structures. The HHR
mapping language can be considered as an alternative
to the FHIR mapping language, that is currently being
specified as part of the FHIR standard. The FHIR
mapping language is an imperative language and
arguably more powerful than the “HHR mapping
language”, but often produces complex descriptions.
Instead the “HHR mapping language” is intended to
be more lightweight. The current prototype of the
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
84
HHR Manager is released on the Artefacts repository
of the CrowdHEALTH project as a jar file named
“hhr-manager-1.3.5.jar”, while the machine-
interpretable definition and mapping of the HHR
model is released as a separate XML file named
“hhr_to_fhir”. The HHR Manager is written in Java
8, while the mapping file is written in XML version
1.0. The HHR Manager generator is released on the
repository of the project as jar file named “hhr-
manager-generator”.
3.2 HHR Manager Library
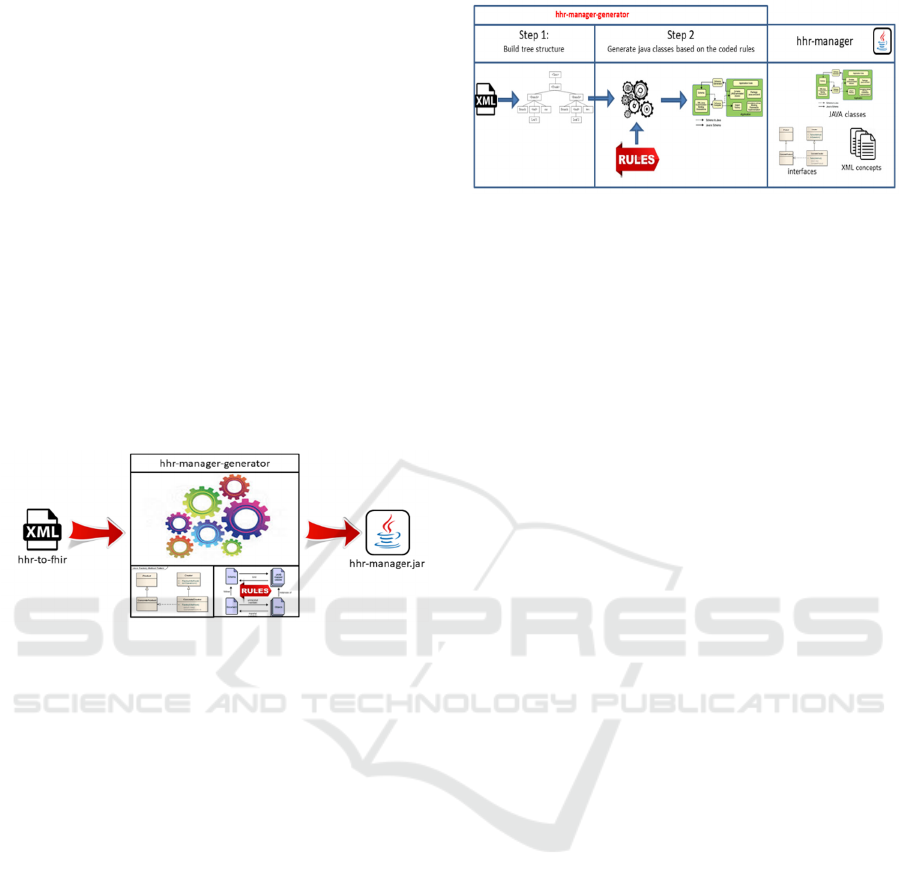
The hhr-manager library is the output of another
developed tool called “hhr-manager-generator”. It
takes as an input the hhr-to-fhir xml file defining the
structure of all classes, attributes and enumerations
included in the HHR model and produces in output
the java code of the hhr manager library (Fig. 3).
Figure 3: The hhr-manager-generator tool.
The hhr-manager-generator tool consists of three
parts:
(i) A set of predefined interfaces to instantiate
and serialize/de-serialize the HHR objects and HHR
concepts (HHRFactory, ConcreteHHRFactory and
Serializer). They are not produced by the tool but they
were written by hands and hard-coded.
(ii) The implementation of a set of rules to
generate the source code (of abstract and concrete
java classes, java interfaces, java enumerations,
attributes, getter and setter methods). There is a set of
rule for each kind of tag of the hhr-maping-language.
(iii) The implementation of a set of rules aiming to
add JAXB annotations to serialize/de-serialize HHR
objects to/from XML documents.
The hhr-manager-generator works in two phases
(Fig. 4): in the first phase a parser analyses the
definition of the HHR model given as input (hhr-to-
fhir.xml) expresses using the hhr mapping language
and builds a hierarchical tree structure. The structure
of the tree is then navigated and the rules for
generation of source code and JAXB annotations are
applied.
Figure 4: Functionality of the hhr-manager-generator.
The output is the generation of the hhr-manager
library containing the source code of Java classes, the
interfaces to instantiate and serialize/de-serialize hhr
objects and standard xml files for the concepts included
in HHR model. Thanks to the hhr-manager-generator
tool it is easy to update the source code of the hhr-
manager whenever new classes, new attributes (or
changes to the existing ones) are added to the HHR
model expressed by the input file hhr-to-fhir.xml.
The introduction of this new tool allows to each
use case (or other stakeholder) to extend the HHR
model by editing the file hhr-to-fhir.xml and generate
the corresponding hhr-manager library. Therefore,
the developer of a use case may choose to use just the
provided XML file describing the final version of the
HHR model or can add it whatever extensions
extension is needed.
Moreover, if some use case requires just to add
new instances to some coded class, then it is not
needed to re-generate the HHR manager. The distinct
files that defined the instances of the <coded> classes
are loaded and interpreted at runtime, therefore the
developer of the use case has just to extend the
content of these files.
3.3 Working Environment
The HHR Manager depends on a standard java virtual
machine that supports Java 8. It can be imported in
any compatible project. Similarly, the mapping file
may be read with any XML parser compatible with
XML version 1.0. Also, the hhr-manager-generator
requires a virtual machine supporting Java 8.
3.4 HHR to FHIR Mapping Example
This section reports an example of usage of the HHR
mapping language. In particular, it shows how to map
the class CarePlan of the HHR model. It maps the
HHR class CarePlan to the homonymous FHIR
resource type CarePlan. An instance of HHR
CarePlan is converted to an instance of FHIR
CarePlan, its attributes type, intent and status are
Holistic Health Records towards Personalized Healthcare
85

mapped respectively to the attributes type, intend and
status of the FHIR resource CarePlan. It should be
noted that, when a class (like CarePlan, in this
example) inherits from a supertype (PersonOrEvent,
in this example), the mapping of all the inherited
attributes may be specified within the definition of the
supertype. If the supertype inherits from another
supertype the mapping is also inherited by the its
supertype and so on. The specification of the mapping
of an HHR class ends where there is a type without
any supertype. In the case of the class CarePlan of the
HHR model, the mapping to FHIR ends in the class
IdentifiedEntity which has not any supertype (the
inheritance chain is IdentifiedEntity,
PersonOrGroupEvent, CarePlan).
The attribute identifier of the HHR class CarePlan
is mapped to the attribute identifier of the FHIR type
CarePlan. The mapping of this attribute is contained
in the tag <class> of the HHR conceptual class
IdentifiedEntity. Note that this HHR conceptual class
has no correspondent FHIR type (indeed the tag-
attribute fhirName is empty). More in details the
value attribute of HHR Identifier is mapped to the
value attribute of FHIR identifier while the attribute
system of the HHR class Identifier is mapped to the
attribute system of the FHIR type Identifier. Note that
isMultipleValue=”true” so there can be more than one
value for the attribute identifier.
The HHR class Agent has no corresponding FHIR
type and therefore the fhirName is set to the empty
string. It is an abstract class having as superclass
IdentifierEntity.
Also, the HHR class PersonOrGroupEvent has no
corresponding FHIR type and its attributes are
mapped within the tags <class> of the subclasses.
The attribute performer of HHR abstract type
Agent is mapped to the attribute performer.actor of
the target FHIR resource type. Note that
isMultipleValue = ”true” so there can be more than
one instance of the attribute performer. The attribute
subject (of HHR abstract type PersonOrGroup) is
mapped to the FHIR attribute subject of the target
resource. The attribute note is mapped to the FHIR
attribute comment. The remaining attributes are
mapped to FHIR extensions of the target FHIR
resource, each one having a specific
StructureDefinition.
asserter (of type HHR abstract class
HealthCarePerson) which StructureDefinition is
defined at http://hl7.org/fhir/StructureDefinition/
asserter
isAutomatic (of HHR type boolean) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/is-automatic
performedWhen (of HHR type Period) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/performed-when
assertedWhen (of HHR type dateTime) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/asserted-when
recorder (of HHR type Agent) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/recorder
recordedWhen (of HHR type dateTime) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/recorded-when
subjectAge (of HHR type depending on the
kind of value set in subjectAge) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/subject-age
duration (of HHR type Duration) which
StructureDefinition is defined at http://hl7.org/fhir/
StructureDefinition/duration
The player of the role class PersonInTime is of
type Person. PersonIntime is an abstract class
inheriting from IdentifiedEntity that has not
corresponding FHIR type. The role class
HealthCarePerson is an abstract and that inherits from
PersonInTime role class. The class CarePlan is
mapped to the homonymous FHIR type CarePlan.
The HHR attribute type is an instance of
CarePlanType and it is mapped to the attribute
category of the target FHIR type CarePlan. The HHR
attribute intent is a mandatory attribute mapped to the
FHIR attribute intent and it can be set with any of the
instances of the CarePlanIntent enum. Finally, the
HHR attribute status is a mandatory inherited
attribute mapped to the FHIR attribute status and it
can be set with any of the instances of the
CarePlanStatus enum. In Fig. 5, it is provided the
final content of the mapping of the CarePlan class of
the HHR model, based on the current description.
4 DISCUSSION
In general, in a context with several sources of data
like the one targeted from the CrowdHEALTH
project, the setting of a baseline allowing the
aggregation of information avoiding ambiguities is
crucial. Many standards and best practices have been
defined over the years with this purpose. Among
them, HL7 FHIR is the specification more tailored to
the needs of the current research. It has been selected
as a ground base for the HHR model, because of its
high coverage of clinical data actually present in the
use cases datasets and of its flexible extension
mechanism that allows the modelling of not yet
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
86

<hhr-to-fhir>
<class hhrName="IdentifiedEntity" fhirName="" isAbstract="true" >
<attribute hhrPath="identifier" hhrType="Identifier" isMultipleValue="true" />
</class>
<class hhrName="Identifier">
<attribute hhrPath="value" hhrType="String" />
<attribute hhrPath="system" hhrType="IdentifierSystem" />
</class>
<class hhrName="Agent" fhirName="" isAbstract="true" hhrSupertype="IdentifiedEntity"/>
<class hhrName="PersonOrGroupEvent" hhrSupertype="IdentifiedEntity" isAbstract="true" fhirName="" >
<attribute hhrPath="performer" hhrType="Agent" isMultipleValue="true" isAbstract="true" fhirPath="performer.actor" />
<attribute hhrPath="asserter" hhrType="HealthCarePerson" isAbstract="true"
fhirExtension="http://hl7.org/fhir/StructureDefinition/asserter" />
<attribute hhrPath="isAutomatic" hhrType="boolean" fhirExtension="http://hl7.org/fhir/StructureDefinition/is-automatic" />
<attribute hhrPath="subject" hhrType="PersonOrGroup" isAbstract="true" />
<attribute hhrPath="performedWhen" hhrType="Period"
fhirExtension="http://hl7.org/fhir/StructureDefinition/performed-when" />
<attribute hhrPath="assertedWhen" hhrType="dateTime" fhirExtension="http://hl7.org/fhir/StructureDefinition/asser-when" />
<attribute hhrPath="recorder" hhrType="Agent" fhirExtension="http://hl7.org/fhir/StructureDefinition/recorder" />
<attribute hhrPath="recordedWhen" hhrType="dateTime" fhirExtension="http://hl7.org/fhir/StructureDefini/recorded-when" />
<attribute hhrPath="subjectAge" hhrType="Range|Duration" isAbstract="true"
fhirExtension="http://hl7.org/fhir/StructureDefinition/subject-age" />
<attribute hhrPath="duration" hhrType="Duration" fhirExtension="http://hl7.org/fhir/StructureDefinition/duration" />
<attribute hhrPath="status" hhrType="EventStatus" isAbstract="true" />
<attribute hhrPath="note" hhrType="String" fhirPath="comment" />
</class>
<role hhrName="PersonInTime" fhirName="" hhrPlayer="Person" hhrSupertype="IdentifiedEntity" isAbstract="true" />
<role hhrName="HealthCarePerson" fhirName="" hhrSupertype="PersonInTime" isAbstract="true"/>
<class hhrName="CarePlan" hhrSupertype="PersonOrGroupEvent" fhirName="CarePlan" >
<attribute hhrPath="type" hhrType="CarePlanType" fhirPath="category" />
<attribute hhrPath="intent" hhrType="CarePlanIntent" fhirPath="intent" fhirMandatory="true" />
<attribute hhrPath="status" hhrType="CarePlanStatus" isInherited="true" fhirPath="status" fhirMandatory="true" />
</class>
<coded hhrName="CarePlanType" fhirName="CodeableConcept" />
<enum hhrName="CarePlanStatus" hhrSupertype="EventStatus" fhirName="code" fhirCodingSystem="http://www.crowdhealth.eu/hhr-
t/consultation-or-treatment-encounter-type">
<instance hhrName="PENDING" fhirCode="pending" fhirCodeDisplay="Pending" />
<instance hhrName="ACTIVE" fhirCode="active" fhirCodeDisplay="Active" />
<instance hhrName="SUSPENDED" fhirCode="suspended" fhirCodeDisplay="Suspended" />
<instance hhrName="COMPLETED" fhirCode="completed" fhirCodeDisplay="Completed" />
<instance hhrName="CANCELLED" fhirCode="cancelled" fhirCodeDisplay="Cancelled" />
</enum>
<enum hhrName="CarePlanIntent" fhirName="code" fhirValueSet="care-plan-intent">
<instance hhrName="PROPOSAL" fhirCode="proposal" fhirCodeDisplay="Proposal" />
<instance hhrName="PLAN" fhirCode="plan" fhirCodeDisplay="Plan" />
<instance hhrName="ORDER" fhirCode="order" fhirCodeDisplay="Order" />
<instance hhrName="OPTION" fhirCode="option" fhirCodeDisplay="Option" />
</enum>
</hhr-to-fhir>
Figure 5: Mapping of the CarePlan class of the HHR Model.
supported clinical data types. FHIR covers a big
number of requirements for representing and
exchanging clinical data, some of them matching with
the CrowdHEALTH requirements, like for example
the modelling of medical observations and clinical
conditions. However, FHIR has some drawback when
used for data integration and analyses. It allows to
represent the same data using different Resource
types, with the risk to produce heterogeneous
representations not easy to aggregate and analyse.
Moreover, it hides important conceptual distinction
that rely on the choice of the right semantic codes.
Therefore, in actual applications, the standard needs
to be constrained to simplify the interoperability. On
the other hand, FHIR does not cover some of the
requirements of the project, lacking a specific
representation of information that is present in the
analysed use cases dataset. For these reasons, a new
model, the HHR model, has been designed and
tailored to the CrowdHEALTH use cases. It
represents information about persons and their
individual roles, the organizations to which the role
players belong, diagnosis and clinical findings of the
patients, medical procedures, medication applications
and related medication and substances administered
to patients, episodes of care and medical encounters
(hospitalization, outpatient, emergency,
hospitalization at home), measurement of vital signs,
physiological parameters, nutritional information,
physical activities results and laboratory test results.
The HHR model has been mapped to FHIR in
order to exploit FHIR as a starting model and to give
the possibility to offer the integrated data using a
standard API. The FHIR extension mechanisms of
FHIR has been used to represent information required
by use cases and modelled in the HHR model, but not
yet present in the FHIR resource. The defined
extensions aim to add details to health-related events,
like the specification of who assert and/or perform an
event during an episode of care and when it occurs,
indicating if the performer is an automatic agent, the
age (or range of age) of the subject at the time the
event occurs, the date when a person is registered into
the system.
Holistic Health Records towards Personalized Healthcare
87

As FHIR also requires the usage of suitable coding
systems, whenever possible standard coding systems
such as ICD-10 and LOINC have been adopted. The
possibility to also use SNOMED CT for encoding
clinical concepts has been investigated. Given the
limitations imposed by its terms of license, only ICD-
10 and LOINC has been adopted as standard
terminologies and any other needed concept not
covered by these terminologies has been represented
with a new terminology defined by CrowdHEALTH.
By maintaining a double view (i.e. conceptual and
logical), the HHR model aims on one hand to
guarantee the interoperability and the possibility to
implement it on top of existing FHIR libraries, and on
the other hand it is also intended to be usable
independently from FHIR (and its future evolutions)
and applicable also for different purposes than only
exchanging health data. For example, it can be more
suitable than FHIR as data schema for Object
Oriented local APIs.
To conclude, CrowdHEALTH healthcare system
integrates a wide set of mechanisms enabling data
acquisition from different sources, cleaning,
aggregation, and transformation into structures that
capture all health determinants, the so-called HHRs.
These HHRs reflect currently 2 million records and
700,000 streams of everyday activities, obtained from
more than 200,000 users, while the system is
expected to exploit the current 75 million
measurements from 1 million people. It is within our
future goals to continuously update this object-
oriented model equivalent to a FHIR profile
expressed both in a human oriented format and in a
machine-oriented format, for supporting additional
data entities, and finally representing more kinds of
information, including social activities and lifestyle
information, as well as real-time workout or daily
activities (CrowdHEALTH D3.1, 2021)
(CrowdHEALTH D3.3, 2021).
ACKNOWLEDGEMENTS
The research leading to the results presented in this
paper has received funding from the European
Union’s funded projects iHELP under Grant
Agreement no 101017441 and CrowdHEALTH
under Grant Agreement no 727560.
REFERENCES
Mavrogiorgou, A., Kiourtis, A., Kyriazis, D. (2017). Plug
‘n’play IoT devices: an approach for dynamic data
acquisition from unknown heterogeneous devices. In
Proc. Of Conference on Complex, Intelligent, and
Software Intensive Systems, pp. 885-895. SPRINGER.
Kiourtis, A., Nifakos, S., Mavrogiorgou, A., Kyriazis, D.
(2019). Aggregating the syntactic and semantic
similarity of healthcare data towards their
transformation to HL7 FHIR through ontology
matching. In International Journal of Medical
Informatics, vol. 132. ELSEVIER.
Geßner, S., et al. (2017). The Portal of Medical Data
Models: Where Have We Been and Where Are We
Going?. In Studies in health technology and
informatics, pp. 858-862. IOS Press.
openEHR, https://www.openehr.org/
HL7 FHIR, https://www.hl7.org/fhir/
LOINC, https://loinc.org/
SNOMED CT, http://www.snomed.org/
ICD-10 Version: 2016, https://icd.who.int/browse10/
2016/en
Kyriazis, D., et al. (2017) CrowdHEALTH: Holistic Health
Records and Big Data Analytics for Health Policy
Making and Personalized Health. In Stud Health
Technol Inform, pp. 19-23. IOS Press.
Monsen, K. A., et al. (2019). Documentation of social
determinants in electronic health records with and
without standardized terminologies: A comparative
study. In Proc. of Singapore Healthcare, pp. 39-47.
SAGE.
ICD-9 Data, http://www.icd9data.com/
What is CPT, https://www.aapc.com/resources/medical-
coding/cpt.aspx
Smith, B., et al. (2007). The OBO Foundry: coordinated
evolution of ontologies to support biomedical data
integration. In Nature biotechnology, vol. 1251.
Cieza, A., et al. (2002). Linking health-status measurements
to the international classification of functioning,
disability and health. In Journal of Rehabilitation
Medicine, pp. 205-210.
Zhe, HE., Geller, H. (2002). Preliminary analysis of
difficulty of importing pattern-based concepts into the
National Cancer Institute thesaurus. In Studies in health
technology and informatics, pp. 228-389. PMC.
Noy, N. F., et al. (2009). BioPortal: ontologies and
integrated data resources at the click of a mouse,
Nucleic acids research. In Nucleic Acids Research. pp.
170-173. Oxford Academic.
Mossakowski, T., Kutz, O., Codescu, M. (2014). Ontohub:
A semantic repository for heterogeneous ontologies. In
Proc. of DACS. CiteSeer.
Lipscomb, C. E. (2000). Medical subject headings (MeSH).
In Bulletin of the Medical Library Association, vol. 265.
PMC.
Lindberg, C. (1990). The Unified Medical Language
System (UMLS) of the National Library of Medicine.
In Journal (American Medical Record Association), pp.
40-42. PMC.
Fontelo, P., Liu, F., Ackerman, M. (2005). ask MEDLINE:
a free-text, natural language query tool for
MEDLINE/PubMed. In BMC Medical Informatics and
Decision Making, vol. 5. Springer.
ICT4AWE 2021 - 7th International Conference on Information and Communication Technologies for Ageing Well and e-Health
88

Liu, S., et al. (2005). RxNorm: prescription for electronic
drug information exchange. In IT professional, pp. 17-
23. IEEE.
CrowdHEALTH D3.1 - Health Record Structure: Design
and Open Specification v1, https://www.crowdhealth.
eu/sites/default/files/crowdhealth/public/content-files/
deliverables/CrowdHEALTH_D3.1%20_Holistic_Hea
lth_Record_Design_Open_%20Specification%20v1.1.
pdf
CrowdHEALTH D3.3 Health Record Structure: Software
prototype v1, https://www.crowdhealth.eu/sites/default/
files/crowdhealth/public/content-files/deliverables/
CrowdHEALTH_D3.3%20Health%20Record%20Struc
ture%20Software%20prototype%20v1.1.pdf
Holistic Health Records towards Personalized Healthcare
89
