
Survival Analysis Algorithms based on Decision Trees with Weighted
Log-rank Criteria
Iulii Vasilev
a
, Mikhail Petrovskiy
b
and Igor Mashechkin
c
Computer Science Department of Lomonosov Moscow State University, MSU, Vorobjovy Gory, Moscow, Russia
Keywords:
Machine Learning, Survival Analysis, Cox Proportional Hazards, Survival Decision Trees, Weighted Log-
rank Split Criteria, Bagging and Boosting Ensembles.
Abstract:
Survival Analysis is an important tool to predict time-to-event in many applications, including but not limited
to medicine, insurance, manufacturing and others. The state-of-the-art statistical approach is based on Cox
proportional hazards. Though, from a practical point of view, it has several important disadvantages, such as
strong assumptions on proportional over time hazard functions and linear relationship between time indepen-
dent covariates and the log hazard. Another technical issue is an inability to deal with missing data directly.
To overcome these disadvantages machine learning survival models based on recursive partitioning approach
have been developed recently. In this paper, we propose a new survival decision tree model that uses weighted
log-rank split criteria. Unlike traditional log-rank criteria the weighted ones allow to give different priority to
events with different time stamps. It works with missing data directly while searching the best splitting point,
its size is controlled by p-value threshold with Bonferroni adjustment and quantile based discretization is used
to decrease the number of potential candidates for splitting points. Also, we investigate how to improve the
accuracy of the model with bagging ensemble of the proposed decision tree models. We introduce an experi-
mental comparison of the proposed methods against Cox proportional risk regression and existing tree-based
survival models and their ensembles. According to the obtained experimental results, the proposed methods
show better performance on several benchmark public medical datasets in terms of Concordance index and
Integrated Brier Score metrics.
1 INTRODUCTION
Survival analysis is a set of statistical models and
methods used for estimating time until the occurance
of an event (or the probability that an event has not
occured). These methods are widely used in demog-
raphy, e.g. for estimating lifespan or age at the first
childbirth, in healthcare, e.g. for estimating duration
of staying in a hospital or survival time after the diag-
nosis of a disease, in engineering (for reliability anal-
ysis), in insurance, economics, and social sciences.
Statistical methods need data, but complete data
may not be available, i.e. the exact time of the event
may be unknown for certain reasons (the event did
not occur before the end of the study or it is unknown
whether it occured). In this case, events are called
censored. The data are censored from below (left cen-
sored) when below a given value the exact values of
a
https://orcid.org/0000-0001-9210-5544
b
https://orcid.org/0000-0002-1236-398X
c
https://orcid.org/0000-0002-9837-585X
observations is unknown. Right censored data (cen-
sored from above) does not have exact observations
above a given value. Further in this paper, right cen-
soring is considered.
The problems studied with the help of survival
analysis are formulated in terms of survival function
(that is compementary distribution function)
S(t) = P(T > t),
where t is observation time and T is random variable
standing for event time. The distribution of T may
also be characterized with so called hazard function
h(t) = −
∂
∂t
logS(t).
There are several ways for estimating the survival
function. A parametric model assumes a distribu-
tion function, and its parameters are estimated based
on the available data. Also we may find empirical
distribution function and then use its complement as
the survival function. Nonparametric methods called
the Kaplan-Meier estimator (Kaplan and Meier, 1958)
132
Vasilev, I., Petrovskiy, M. and Mashechkin, I.
Survival Analysis Algorithms based on Decision Trees with Weighted Log-rank Criteria.
DOI: 10.5220/0010987100003122
In Proceedings of the 11th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2022), pages 132-140
ISBN: 978-989-758-549-4; ISSN: 2184-4313
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

and Nelson-Aalen (Nelson, 1972) estimator are more
powerful. The Kaplan-Meier estimator has the form
S(t) =
∏
i:t
i
≤t
1 −
d
i
n
i
,
where t
i
is time of the event, d
i
is number of events
that occurred at time t
i
, and n
i
number of events after
time t
i
(or unknown at t
i
). Nelson-Aalen esimator ap-
plies the same idea to the cumulative harazd function
H(t) =
R
t
0
h(s)ds and then transforms it to the estima-
tion of the survival function.
In real-life problems, especially in studing dis-
eases and mortality, data may contain covariate infor-
mation (gender, age etc.), and the question is how it
affects the survival function. Let X be a random vec-
tor of covariates and T be a non-negative random sur-
vival time. For an observation with a covariate vector
x, we determine the probability that an event occur
later than a certain time t ≥ 0 as a conditional survival
function
S(t | x) = P(T > t | X = x).
The correstonding conditional hazard function is
h(t | x) = −
∂
∂t
logS(t | x).
The Cox proportional hazards model (Cox, 1972)
is one of the most popular models for taking covari-
ates into account. The model is based on the assump-
tion that all observations have the same form of the
conditional hazard function:
h(t | x) = h
0
(t)exp
x
T
β
,
where h
0
(t) is baseline hazard function, x is a vector
of covariates, and β is a vector of weights for each co-
variate. The corresponding conditional survival func-
tion
S(t | x) = S
0
(t)
exp
(
x
T
β
)
,
may be predicted for a particular observation with the
use of the Breslow estimate (Lin, 2007) for the base-
line survival function S
0
(t) and weights β.
However, the method has several significant dis-
advantages:
• The ratio of hazard functions for two different
vector is constant over time.
• Significance of covariates does not change with
time. In clinical practice, the influence of factors
on risk can vary over time. For example, a patient
is more at risk after surgery and more stable after
rehabilitation.
• Linear combination of covariates may have no
ground for the particular set of covariates.
When we faced a real-life problem, we realized
that the above mentioned approaches couldn’t be ap-
plied due to the following reasons. First of all,
they are not sensitive to the specificity of datasets,
in particular earlier events have the same affect to
the estimation then the later ones. Second, they do
not deal with missing values often presented in real
datasets. Therefore we turn to the tree-based ap-
proaches and develop new method devoid of the listed
drawbacks. Experimental results on real datasets
show that the proposed approach outperforms other
tree-based methods and the Cox model as well.
The paper is organized as follows: in section 2,
we review the most popular machine learning meth-
ods for survival analysis: survival tree, random sur-
vival forest and gradient boosting survival analysis.
Based on weighted logrank criteria, we propose a new
approach for constructing decision trees and their en-
sembles in section 3. Section 4 is devoted to the re-
sults of an experimental study, they compare the con-
sidered methods on two public benchmark datasets
from healthcare area. In section 5, we present the
main results of the paper and future research direc-
tions.
2 OVERVIEW
To solve the problem of survival analysis, the source
data can be presented as three groups of features: in-
put features X (covariates) at the time of the study,
time T from the beginning of study to event occur-
rence, binary indicator of the event occurrence E (ob-
servations with E = 0 will be considered censored).
To solve the problem of predicting the survival
function for new data, a model of the form M(X) =
ˆ
S(t), where
ˆ
S(t) is an estimate of the survival function
S(t) constructed from the target data T and E, can be
built on the available inpute data X.
In this section, various approaches for building
prediction models M(X) are discussed, in particu-
lar: Survival Tree (LeBlanc and Crowley, 1993), Ran-
dom Survival Forest (Ishwaran et al., 2008), Gradient
Boosting Survival Analysis (Friedman, 2001).
Tree-based methods are based on the idea of re-
cursive partitioning the feature space to groups (de-
scribed by nodes) similar according to a split crite-
rion. This idea was firstly introduced by Morgan and
Sonquist (Morgan and Sonquist, 1963). All of the
observations are placed at the root node, and then
the best of possible binary splits is chosen in accor-
dance with the predefined criterion. The process is re-
peated recursively on the children nodes until a stop-
ping condition is satisfied. The tree for big dataset
Survival Analysis Algorithms based on Decision Trees with Weighted Log-rank Criteria
133

is usually very large, and a pruning method is to be
applied. Trees ensembels lead to smaller trees and
help to avoid the problem of the best tree selection
and overfitting.
2.1 Survival Tree
Ciampi et al. (Ciampi et al., 1986) suggested to use
the logrank statistic (Lee, 2021) for comparison of the
two groups of observation in the children nodes. The
more is the value of the statistic the more the hazarad
functions of the groups differ. The splitting is chosen
for the largest statistic value. Leblanc M. and Crow-
ley J. (LeBlanc and Crowley, 1993) introduced a tree
algorithm based on logrank statistic in combination
with the cost-complexity pruning algorithm.
Suppose the observations are divided into two
groups somehow. For the two groups, define an or-
dered set of event times: τ
1
< τ
2
< ... < τ
K
. Let
N
1, j
and N
2, j
be the number of subjects at time τ
j
(under observation or censored), and O
1, j
and O
2, j
be the observed number of events at time τ
j
. Then
the total numbers at time τ
j
are N
j
= N
1, j
+ N
2, j
and
O
j
= O
1, j
+ O
2, j
. The expected number of events at
τ
j
is E
i, j
=
N
i, j
O
j
N
j
. Based on the available data, we can
calculate the weighted logrank statistic:
LR =
∑
K
j=1
w
j
(O
1, j
− E
1, j
)
r
∑
K
j=1
w
2
j
E
1, j
N
j
−O
j
N
j
N
j
−N
1, j
N
j
−1
. (1)
The weights are chosen for correcting the influence of
early or late events.
The following values control the tree growth:
maximum tree depth, maximum number of covatiates
for searching the best partition, maximum number of
tree leaves and minimum number of observations in a
node.
2.2 Random Survival Forest
The Random Survival Forest model proposed in (Ish-
waran et al., 2008) and based on the idea of construct-
ing an ensemble of survival trees (LeBlanc and Crow-
ley, 1993) and aggregating their predictions:
1. Constructs N bootstrap samples (with resampling)
from the source data. Each bootstrap subsample
excludes 37% of the data on average, the excluded
data is called out-of-bag (OOB) sample.
2. On each bootstrap sample, a survival tree is con-
structed. The splitting at each node of the tree is
based on P randomly selected covariates. The best
partition maximizes the difference between chil-
dren nodes (in particular, measured with logrank
statistic) is chosen.
3. Survival trees are constructed until the bootstrap
sample is exhausted.
For the constructed ensemble, we can calculate
the prediction error based on the out-of-bag data
OOB
i
, i = 1...N. For an observation from the origi-
nal sample with a covariate vector x, the prediction is
the average prediction over the trees with x ∈ OOB
i
.
The prediction of the survival function for an ob-
servation with a covariate vector x is calculated as the
average prediction over all trees in the ensemble for
all time points. The survival tree prediction is the
Kaplan-Meier estimate calculated for the data asso-
ciated with the same leaf as x. Averaging the decision
tree predictions improves accuracy and avoids over-
fitting.
The following parameters are to be chosen when
constructing an ensemble: number of trees in the en-
semble N, bootstrap sample size, single tree growth
control parameters, the number of randomly choosen
covariates for each split search.
2.3 Gradient Boosting Survival Analysis
Another popular approach for constructing an ensem-
ble of trees is Gradient Boosting introduced by Fried-
man and Jerome H. (Friedman, 2001). Unlike Ran-
dom Survival Forest based on independent tree con-
structing and averaging their predictions, the Gradient
Boosting Survival Analysis algorithm (Hothorn et al.,
2006) uses an iterative tree learning. Aggregation of
tree forecasts is made with weighting coefficients cal-
culated when a new tree is added to the ensemble.
The purpose of the Gradient Boosting Survival
Analysis algorithm is to minimize the loss-function
L(y, F(x)) that defines the ensemble error. In the sur-
vival analysis, the loss function is usually calculated
as the deviation from the logarithmic Cox partial like-
lihood function (Cox, 1972). Let
{
(x
i
, y
i
)
}
n
i=1
be the
training set, L be the loss function, M be the ensem-
ble size. The general algorithm of Gradient Boosting
Survival Analysis has the following steps:
1. Initialize model with a constant value α such as::
F
0
(x) = argmin
α
n
∑
i=1
L(y
i
, α)
2. For m = 1 to M:
(a) Compute pseudo-residuals for i = 1,...,n:
r
im
= −
∂L(y
i
, F(x
i
))
∂F(x
i
)
F(x)=F
m−1
(x)
(b) Fit a survival tree h
m
(x) using the training set
{
(x
i
, r
im
)
}
n
i=1
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
134

(c) Compute weight v
m
(0 < v
m
< 1) of survival
tree by solving the following optimization
problem:
v
m
= argmin
v
n
∑
i=1
L(y
i
, F
m−1
(x
i
) + v · h
m
(x
i
))
(d) Update the ensemble:
F
m
(x) = F
m−1
(x) + v
m
· h
m
(x),
3. The final ensemble is F
M
The prediction of the survival function for an ob-
servation with a covariate x is computed as a weighted
sum of predictions of all tree in the ensemble for all
points in time.
The following parameters should be chosen when
constructing the Gradient Boosting Survival Analy-
sis: loss function L, ensemble size M, method of cal-
culating the weights of trees v
m
and parameters for
single tree growth control.
3 PROPOSED APPROACH
Several significant problems arise when the known
methods are applied to the real datasets. The exist-
ing tree approaches 2.1, 2.2 use the logrank criterion
(1) for evaluating the difference of samples to find
the best split. The first problem is that the criterion
is calculated under the assumptions that the censor-
ing indicator is uncorrelated with prediction and sur-
vival probabilities are the same for events in early and
late stages of the study. Researches presented in (Lee,
2021), (Buyske et al., 2000) suggest to use a weight
function incorporated in the logrank to improve its
sensitivity.
Second, the existing approaches usually work
with fully complete data. In practice, the problem of
missing values is very common, and the reason for
the appearance may be unknown, it means that proper
imputation method is hard to find. To apply the mod-
els to real dataset, an approach for handling missing
values must be developed.
The problem of applicability of algorithms for
large amounts of data is also very important. In such
data, the initial assumptions of the models may be
violated, and the complexity of the computation in-
creases. Although the hyperparameters of the model
may bound the complexity of existing approaches,
this may affect the prediction accuracy. The problem
of increasing complexity may be solved by incorpo-
rating additional principles of feature processing with
the similar complexity for any volume of data.
3.1 Weighted Log-rank Criteria
To solve the problem of low sensitivity of the logrank
criterion to early events, it is proposed to investigate
the applicability of weighted logrank criteria such as
Wilcoxon (Breslow, 1970), Tarone-Ware (Tarone and
Ware, 1977), Peto-Peto (Peto and Peto, 1972) tests.
In the task of building tree based survival models, the
applicability of weighted criteria has not been inves-
tigated previously.
In general, the weighted criteria are based on de-
termining the weights w
j
in (1):
1. generalized Wilcoxon criterion: w
j
= N
j
.
The statistics are constructed by weighting the
contributions with the number of observations at
risk. It assigns greater weights for early events for
larger number of observations. However, this cri-
terion depends a lot on difference in the censoring
structure of the groups.
2. Peto-Peto criterion: w
j
=
ˆ
S(τ
j
), where
ˆ
S(t) is the
Kaplan-Meier estimator for the survival function.
The criterion is suitable for cases with dispro-
portionate hazard functions. However, unlike the
Wilcoxon test, differences in the censoring struc-
ture do not affect the criterion.
3. Tarone-Ware criterion: w
j
=
p
N
j
.
The statistic is constructed by weighting the con-
tribution by the square root of the number of
observations at risk. Like the Wilcoxon crite-
rion, it assigns higher weights (though not so
large) to earlier events. The study (Klein and
Moeschberger, 1997) notes that the criterion is the
“golden mean” between the Wilcoxon and Peto-
Peto criteria.
3.2 Proposed Decision Tree
In this paper, we propose the following approach for
constructing a survival tree. As in 2.1, 2.2, we start
with the root node containing all observations. Each
node is partitioned recursively into two child nodes
according the best value of a splitting criterion. Con-
sider the algorithm of finding the best split in an ran-
dom node ND based on the specified set of features
F
ND
:
1. For each feature f ∈ F
ND
:
(a) If f is a continuous feature:
i. Intermediate points a
1
, a
2
, ...a
k
by unique val-
ues v
i
of the feature f : v
1
< a
1
< v
2
<
a
2
...a
n−1
< v
n
.
ii. Let’s limit the maximum number of interme-
diate points to the number k. If n > k, then the
Survival Analysis Algorithms based on Decision Trees with Weighted Log-rank Criteria
135
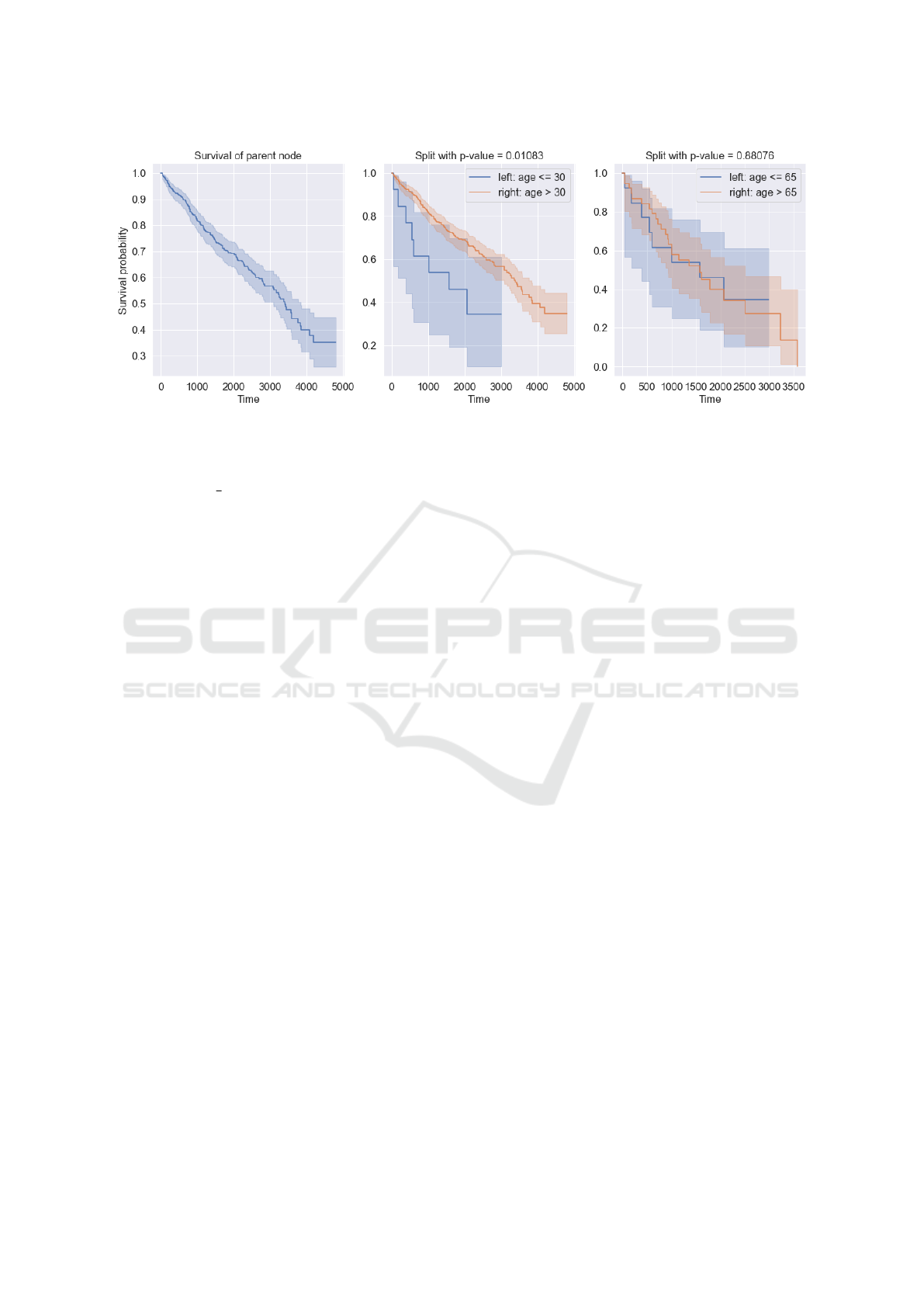
Figure 1: Example of two splits of a parent node based on age and values 30 and 65 with p-value for each split. A split with a
value of 30 has a smaller p-value and defines more different child nodes.
values of the functions are discretized, and the
quantile
i
k
by f are taken as split points a
i
.
iii. For each split point a
i
, splitting results in two
samples: le f t (with f ≤ a
i
) and right (with
f > a
i
).
(b) If f is a categorical feature:
i. All possible pairs of non-overlapping sets l, r
of unique values of the feature f are consid-
ered for splitting.
ii. For each pair of values l
i
, r
i
, we construct two
samples: le f t (with f ∈ l) and right (with f ∈
r).
(c) Estimate the difference between survival func-
tions on the left and right samples as an upper-
tailed p-value = 1–cdf(z), where z is the value
of the weighted statistics 1 having cumulative
Chi-squared distribution function cdf (Aster
et al., 2018). The lower p-value means the big-
ger difference in survival functions on the left
and right branches.
For example, the Figure 1 shows the survival
function of the parent node and two splits based
on the feature ”age” with values 30 and 65.
When split by 30, the p-value is 0.01 and the
survival functions are different. When parti-
tioned by the value of 65, the p-value is 0.88
and the survival functions are very close. Con-
sequently, to maximize the difference between
child nodes, we choose a split with a smaller
p-value.
(d) Missing values of feature f can be handled by
the following algorithm: the values are added to
each samples le f t and right in turn and the p-
value is calculated. Finally, the missing values
are added to the sample with minimal p-value.
(e) Choose a pair of split le f t, right with a mini-
mum p-value.
2. Choose the best feature in the node:
(a) Apply the Bonferroni adjustment (Benjamini
and Hochberg, 1995) to the selected p-value for
each feature. This adjustment reduces the sig-
nificance of the more common features, giving
preference to the rarer significant splits.
(b) Choose the feature with the minimal p-value by
the best partition le f t, right.
Applying the described approach of splitting node
into child nodes, a decision tree is constructed for
the source data. An example of the constructed de-
cision tree of depth 2 is shown in Figure 2. To control
tree growth, the following parameters are used: max-
imum tree depth, maximum number of features when
searching for the best partitioning, minimum number
of observations in each node, level of partitioning sig-
nificance, maximum number of split points for one
feature.
For an observation with a feature vector x, data in
the same leaf node as x allows to predict probability
and time of event as an aggregation of outcomes and
times based on median, mean, or weighted sum; sur-
vival function: the Kaplan-Meier estimator calculated
on the sample in the leaf node.
3.3 Proposed Ensemble of Decision
Trees
The proposed decision tree approach can be applied
for constructing bagging ensembles of decision trees.
Aggregation of predictions from several models im-
proves accuracy and prevents overfitting.
We propose a bagging approach based on iterative
decision tree ensemble construction:
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
136

Figure 2: Example of a constructed decision tree of depth 2 with visualization of the survival function estimate at each node.
The tree is based on the following features: bili (serum bilirubin), protime (standardized blood clotting time), age.
1. A bootstrap sample of a predefined size (speci-
fied as a hyperparameter) is constructed, all ob-
servations have equal probability to be selected.
The part of observations out of bootstrap sample
is named as OOB.
2. Based on the approach proposed in subsection 3.2,
a decision tree is built on the boostrap sample.
3. Calculates the OOB-error of the ensemble before
and after adding the next decision tree model. The
error is calculated similarly to the RSF 2.2 model:
the prediction for observation x is the aggregation
of tree predictions for which x ∈ OOB
i
. The error
calculation metric is specified as a hyperparame-
ter.
4. If the added model increases the ensemble error,
the model is deleted from ensemble, and the con-
struction is terminated. Otherwise, the algorithm
returns to step 1
5. Also, the algorithm may work in a tolerance
mode: an ensemble of decision trees are sequen-
tially built for a predetermined number of models
N, OOB-error is calculated at each iteration, and
the final number of models in the ensemble is de-
termined by the minimum error over all iterations.
The bagging model prediction is the aggregation
of model predictions in the ensemble (median, mean,
or weighted mean can be used). In particular, the
survival function is computed as an aggregation over
each time point.
To control for computational complexity, the fol-
lowing parameters are used: maximum number of
trees in the ensemble, bootstrap sample size, toler-
ance mode flag, the method of aggregation of ensem-
ble model predictions, metric for calculating OOB er-
ror, parameters of singlr tree growth control.
4 EXPERIMENTS
4.1 Metrics
In this paper, we use Concordance index and In-
tegrated Brier Score metrics to evaluate the perfor-
mance of the proposed prediction models and to com-
pare them with existing ones. The Concordance Index
(Harrell Jr et al., 1996) is widely used in survival anal-
ysis. It is similar to AUC in the sense that it measures
the fraction of concordant or correctly ordered pairs
of samples among all available pairs in the dataset.
The highest value of the metric is one (if the order is
perfect), and the value of 0.5 means that the model
produces completely random predictions.
The following formula is used for calculating the
concordance index:
CI =
∑
i, j
1
T
j
<T
i
· 1
η
j
<η
i
∑
i, j
1
T
j
<T
i
,
Survival Analysis Algorithms based on Decision Trees with Weighted Log-rank Criteria
137

where T
k
is the true time of the event, and η
k
is the
time predicted by the model.
However, this metric is based only on the pre-
dicted time of the event, and it does not allow esti-
mating the survival function. The value of CI does not
change when the survival function is biased, although
the predicted time is highly distorted compared to the
true time.
To eliminate this problem, we use a metric called
Integrated Brier Score (Murphy, 1973), (Brier and
Allen, 1951), (Haider et al., 2020) based on the devi-
ation of the predicted survival function from the true
one (equal to 1 before the event occurs and 0 after
that). The Brier Score (BS) metric (Brier and Allen,
1951) is used for estimating the performance of the
prediction at a fixed time point t and is calculated in
the following way:
BS(t) =
1
N
∑
i
(
(0 − S(t, x
i
))
2
if T
i
≤ t
(1 − S(t, x
i
))
2
if T
i
> t
(2)
where S(t, x
i
) is the prediction of the survival function
at time t for observation x
i
with event time T
i
.
Next, the squares of variance are averaged over
all observations at time t. The best BS value is 0,
in the case the predicted and true survival functions
coincide. However, 2 does not take into account the
censoring. In such a case, the following modification
of the BS (Murphy, 1973),(Haider et al., 2020) can be
used:
BS(t) =
1
N
∑
i
(0−S(t,x
i
))
2
G(T
i
)
if T
i
≤ t, δ
i
= 1
(1−S(t,x
i
))
2
G(t)
if T
i
> t
0 if T
i
= t, δ
i
= 0
(3)
As in (2), S(t, x
i
) is a prediction of the survival
function at time t for observation x
i
with event time T
i
.
The parameter δ
i
in (3) is the censored flag of obser-
vation x
i
, it is equal to 1 if the event occurred and 0 if
the event is censored. The function G(t) = P(c > t) is
the Kaplan-Meier estimation of the survival function
constructed on the censored observations (the cen-
sored flag is reversed when constructing the estimate).
The squares of variance in (3) are adjusted by weight-
ing inverse probability of non-censoring:
1
G(T
i
)
if the
event occurs before t, and
1
G(t)
if the event occurs af-
ter t. Observations censored before t are not used in
calculation.
To aggregate the BS estimates over all time mo-
ments, the Integrated Brier Score is used:
IBS =
1
t
max
t
max
Z
0
BS(t)d t
4.2 Datasets
In the experiments we use the following public medi-
cal benchmark datasets.
The Primary Biliary Cirrhosis (PBC) dataset (Ka-
plan, 1996) was collected in the time period from
1974 to 1984. The death is considered as the event.
The dataset contains 276 observations and 17 features
including cirrhosis status, treatment strategy, and clin-
ical measures. Also, 12 features such as treatment
strategies and clinical indicators may be missed, with
the maximum number of missings in the cholesterol
indicator (134 missings) and in the triglyceride indi-
cator (136 missings). At the end of the study, there
were 263 patients for whom there were no fatal out-
comes.
The German Breast Cancer Study Group (GBSG)
(Schumacher, 1994) dataset was collected in the pe-
riod from 1984 to 1989. The cancer relapse is consid-
ered as the event. The data set contains 686 observa-
tions and 8 features such as tumor characteristics and
treatment strategies. The dataset does not have miss-
ings. At the end of the study there were 387 patients
without relapse.
4.3 Experimental Setup
For honest estimating of the models performance, we
implemented hyperparameters grid search using 5-
folds cross validation (Refaeilzadeh et al., 2009). All
variable hyperparameters and their grid characteris-
tics are presented in the table 1. The best vector of
hyperparameters for the model is selected according
to the minimum value of cross-validated IBS metric.
4.4 Results
For the existing methods we used scikit-survival li-
brary (P
¨
olsterl, 2020) implementation. For the pro-
posed methods we implemented our own code. The
results of performance estimating for all methods by
CI, IBS metrics on PBC and GBSG datasets are pre-
sented in tables 2, 3 (top 3 models by each metric are
marked in bold).
On the PBC dataset the Gradient Boosting Sur-
vival Analysis method showed the best C I, but pro-
posed in the paper Bagging Wilcoxon and Bagging
Tarone-Ware methods are next and close to the leader.
Moreover, on IBS metric proposed Bagging Peto and
Bagging Tarone-Ware methods outperformed others.
That is important, because IBS metric is more ap-
propriate for evaluating the performance of survival
analysis models since it estimates the deviation of the
predicted survival function from the true one. On
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
138
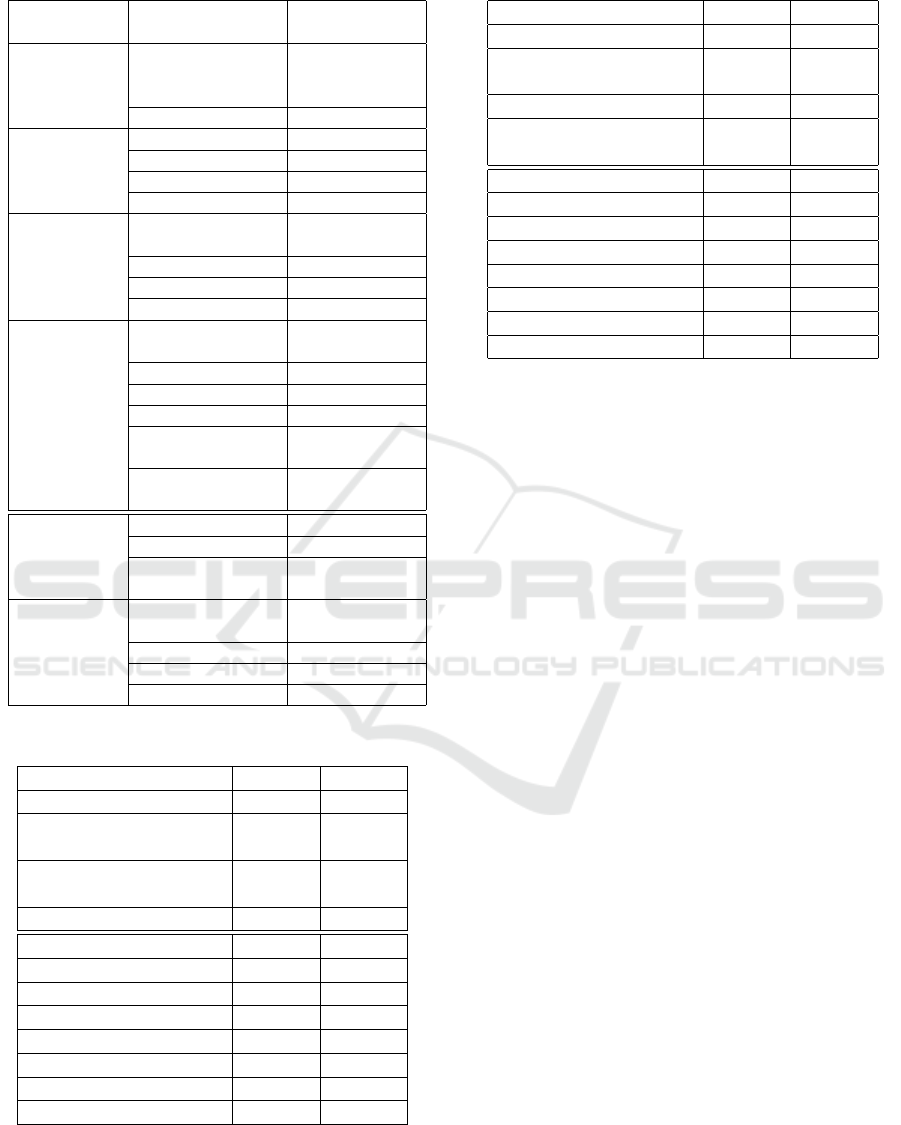
Table 1: Hyperparameters of predictive models.
Predictive
model
Hyperparameter Values
CoxPH Sur-
vival Analy-
sis
regularization
penalty
0.1, 0.01, 0.001
ties breslow, efron
Survival Tree split strategy best, random
max depth 10 to 30 step 5
min sample leaf 1 to 20 step 1
max features sqrt, log2, None
Random Sur-
vival Forest
num estimators 10 to 100 step
10
max depth 10 to 30 step 5
min sample leaf 1 to 20 step 1
max features sqrt, log2, None
Gradient
Boosting SA
num estimators 10 to 100 step
10
max depth 10 to 30 step 5
min sample leaf 1 to 20 step 1
max features sqrt, log2, None
loss function coxph, squared,
ipcwls
learning rate 0.01 to 0.5 step
0.01
Tree max depth 10 to 30 step 5
min sample leaf 1 to 20 step 1
significance
threshold
0.01, 0.05, 0.1,
0.15
Bagging bootstrap sample
size
0.3 to 0.9 step
0.1
num estimators 10 to 50 step 5
max depth 10 to 30 step 5
min sample leaf 1 to 20 step 1
Table 2: PBC dataset results.
Predictive model CI IBS
Survival Tree 0.61325 0.25292
Gradient Boosting Sur-
vival Analysis
0.65536 0.23480
CoxPH Survival Analy-
sis
0.63965 0.23050
Random Survival Forest 0.65060 0.20516
Tree tarone-ware 0.64744 0.26982
Tree wilcoxon 0.64443 0.25092
Tree logrank 0.63582 0.23240
Tree peto 0.64001 0.21770
Bagging wilcoxon 0.65284 0.21341
Bagging logrank 0.63783 0.20829
Bagging peto 0.64988 0.20258
Bagging tarone-ware 0.65118 0.20104
the GBSG dataset all proposed Bagging methods out-
performed their existing competitors by both CI, IBS
metrics.
Table 3: GBSG dataset results.
Predictive model CI IBS
Survival Tree 0.58500 0.19119
Gradient Boosting Sur-
vival Analysis
0.60818 0.17768
Random Survival Forest 0.61795 0.17352
CoxPH Survival Analy-
sis
0.61281 0.17324
Tree logrank 0.58162 0.19082
Tree peto 0.59781 0.18582
Tree wilcoxon 0.59192 0.18510
Tree tarone-ware 0.60029 0.18489
Bagging logrank 0.61861 0.17262
Bagging wilcoxon 0.62707 0.17157
Bagging tarone-ware 0.62276 0.17112
Bagging peto 0.62252 0.17079
Also, it is important to note that in many cases the
proposed single-model decision trees with weighted
long-rank criteria outperform their single-model com-
petitors such as Survival tree and Cox PH regularized
regression.
Consequently, the best prediction models for both
datasets are based on bagging ensembles of decision
trees with weighted log-rank split criteria.
5 CONCLUSIONS
In this paper, we have proposed a method for build-
ing nonlinear survival models based on the recursive
partitioning with weighted log-rank test as a split cri-
terion. This approach allows avoid some disadvan-
tages of the existing state-of-the-art methods in sur-
vival analysis and build survival models that do not
exploit such assumptions as proportionality of haz-
ards over time and linear dependence between the log
of hazard and combination of covariates. Besides, the
proposed method effectively works with missing val-
ues, can pay greater attention to the events with ear-
lier occurrence time. Using Bonferroni adjustment for
weighted log-rank in splitting procedure allows more
correct comparisons among candidate features with
different power. We have also experimentally shown
on medical becnchmark datasets that the bagging en-
sembles of the proposed models outperform the ex-
isting models and their ensembles in terms of the
Concordance index and Integrated Brier Score met-
rics. In further research, we plan to study the behav-
ior of boosting ensembles of the proposed models, to
develop efficient algorithms for time-efficient finding
the optimal split with weighted log-rank criteria for
high-power categorical features and on large dataset,
Survival Analysis Algorithms based on Decision Trees with Weighted Log-rank Criteria
139

as well as to investigate the performance of the pro-
posed methods on other benchmark datasets, includ-
ing cases from other application areas.
REFERENCES
Aster, R. C., Borchers, B., and Thurber, C. H. (2018). Pa-
rameter estimation and inverse problems. Elsevier.
Benjamini, Y. and Hochberg, Y. (1995). Controlling the
false discovery rate: a practical and powerful ap-
proach to multiple testing. Journal of the Royal statis-
tical society: series B (Methodological), 57(1):289–
300.
Breslow, N. (1970). A generalized kruskal-wallis test for
comparing k samples subject to unequal patterns of
censorship. Biometrika, 57(3):579–594.
Brier, G. W. and Allen, R. A. (1951). Verification of weather
forecasts. In Compendium of meteorology, pages 841–
848. Springer.
Buyske, S., Fagerstrom, R., and Ying, Z. (2000). A class
of weighted log-rank tests for survival data when the
event is rare. Journal of the American Statistical As-
sociation, 95(449):249–258.
Ciampi, A., Thiffault, J., Nakache, J.-P., and Asselain, B.
(1986). Stratification by stepwise regression, corre-
spondence analysis and recursive partition: a compar-
ison of three methods of analysis for survival data with
covariates. Computational statistics & data analysis,
4(3):185–204.
Cox, D. R. (1972). Regression models and life-tables. Jour-
nal of the Royal Statistical Society: Series B (Method-
ological), 34(2):187–202.
Friedman, J. H. (2001). Greedy function approximation: a
gradient boosting machine. Annals of statistics, pages
1189–1232.
Haider, H., Hoehn, B., Davis, S., and Greiner, R. (2020). Ef-
fective ways to build and evaluate individual survival
distributions. J. Mach. Learn. Res., 21:85–1.
Harrell Jr, F. E., Lee, K. L., and Mark, D. B. (1996).
Multivariable prognostic models: issues in develop-
ing models, evaluating assumptions and adequacy, and
measuring and reducing errors. Statistics in medicine,
15(4):361–387.
Hothorn, T., B
¨
uhlmann, P., Dudoit, S., Molinaro, A., and
Van Der Laan, M. J. (2006). Survival ensembles. Bio-
statistics, 7(3):355–373.
Ishwaran, H., Kogalur, U. B., Blackstone, E. H., and Lauer,
M. S. (2008). Random survival forests. The annals of
applied statistics, 2(3):841–860.
Kaplan, E. L. and Meier, P. (1958). Nonparametric esti-
mation from incomplete observations. Journal of the
American statistical association, 53(282):457–481.
Kaplan, M. M. (1996). Primary biliary cirrhosis. New Eng-
land Journal of Medicine, 335(21):1570–1580.
Klein, J. P. and Moeschberger, M. L. (1997). Statistics
for biology and health. Stat. Biol. Health, New York,
27238.
LeBlanc, M. and Crowley, J. (1993). Survival trees by good-
ness of split. Journal of the American Statistical As-
sociation, 88(422):457–467.
Lee, S.-H. (2021). Weighted log-rank statistics for acceler-
ated failure time model. Stats, 4(2):348–358.
Lin, D. (2007). On the breslow estimator. Lifetime data
analysis, 13(4):471–480.
Morgan, J. N. and Sonquist, J. A. (1963). Problems in the
analysis of survey data, and a proposal. Journal of the
American statistical association, 58(302):415–434.
Murphy, A. H. (1973). A new vector partition of the proba-
bility score. Journal of Applied Meteorology and Cli-
matology, 12(4):595–600.
Nelson, W. (1972). Theory and applications of hazard
plotting for censored failure data. Technometrics,
14(4):945–966.
Peto, R. and Peto, J. (1972). Asymptotically efficient rank
invariant test procedures. Journal of the Royal Statis-
tical Society: Series A (General), 135(2):185–198.
P
¨
olsterl, S. (2020). scikit-survival: A library for time-to-
event analysis built on top of scikit-learn. J. Mach.
Learn. Res., 21(212):1–6.
Refaeilzadeh, P., Tang, L., and Liu, H. (2009). Cross-
validation. Encyclopedia of database systems, 5:532–
538.
Schumacher, M. (1994). Rauschecker for the german breast
cancer study group, randomized 2× 2 trial evaluating
hormonal treatment and the duration of chemotherapy
in node-positive lbreast cancer patients. Journal of
Clinical Oncology, 12:2086–2093.
Tarone, R. E. and Ware, J. (1977). On distribution-free
tests for equality of survival distributions. Biometrika,
64(1):156–160.
ICPRAM 2022 - 11th International Conference on Pattern Recognition Applications and Methods
140
