
Analysis of Dominant Spoilage Bacteria in Beijing Sausages
Wenhui Liang
1,
†
a
, Fang Wang
1,
†
b
, Ting Li
1c
, Jiayun Kang
1d
, Yu Hao
1e
, Siyu Shi
1f
and Jinghua Qi
1,2 g
1
Beijing Key Laboratory of Agricultural, Product Detection and Control of Spoilage Organisms and Pesticide Residue,
Beijing University of Agriculture, Beijing, China
2
Beijing Innovation Consortium of Swine Research System, Beijing, China
f
2816155579@qq.com,
g,*
abc960718@sina.com
*
Corresponding author
†
Contributed equally to this work
Keywords:
Beijing Sausages, Isolation, Identification, Spoilage Bacteria.
Abstract:
Low temperature-heated meat products, which are popular among consumers because of their unique flavor,
texture, and nutrition, have a relatively short shelf life due to the low sterilization temperature and are therefore
prone to spoilage. The profile of bacteria associated with spoilage of Beijing sausages is here summarized
through isolation, identification and spoilage microflora analysis. The main spoilage bacteria isolated and
identified from Beijing sausages were one genus of Pseudomonas sp., one genus of Staphylococcus sp. and
two genera of Brochothrix sp. By denaturing gradient gel electrophoresis (PCR-DGGE) analysis, we
identified 13 spoilage bacteria of Beijing sausages, of which Pseudomonas sp. was the most abundant.
1 INTRODUCTION
1
Beijing sausage is a low-temperature meat product
which has the advantages of simple production
technology, low sterilization temperature, slight
protein denaturation, compact meat quality, elasticity,
and delicious taste. However, due to the
characteristics of the processing technology, the
heating temperature of Beijing sausages fails to
effectively sterilize the product, and the nutrition
profile provides good conditions for the growth of
microorganisms. Thus, Beijing sausages are prone to
spoilage during storage, transportation, and sale, so
the shelf life of these products is often short, greatly
limiting the development of such products.
The key to extending the shelf life of meat
products is to study the species and characteristics of
the dominant bacteria involved in spoilage, and then
a
https://orcid.org/0000-0001-8982-7693
b
https://orcid.org/0000-0002-0438-6786
c
https://orcid.org/0000-0002-5644-5768
d
https://orcid.org/0000-0003-0174-9710
e
https://orcid.org/0000-0001-9311-3562
f
https://orcid.org/0000-0002-2461-4643
g
https://orcid.org/0000-0001-9204-3697
select effective preservation measures. In recent
years, the species and characteristics of spoilage
bacteria in some meat products have been studied in
domestic and foreign literature (Adams et al., 2007;
Danilo et al., 2009; Doulgeraki et al., 2012; Liu et al.,
2010; Peirson et al., 2003), collectively showing that
the types of dominant bacteria involved in spoilage
vary between meat products. Here, the bacterial
distribution after spoilage in Beijing sausages was
analyzed. The dominant bacteria causing the spoilage
were isolated, purified, and identified to provide the
theoretical basis for taking effective anti-spoilage
measures and thus extending the shelf life of Beijing
sausages.
84
Liang, W., Wang, F., Li, T., Kang, J., Hao, Y., Shi, S. and Qi, J.
Analysis of Dominant Spoilage Bacteria in Beijing Sausages.
DOI: 10.5220/0011182900003443
In Proceedings of the 4th International Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 84-88
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 MATERIALS AND METHODS
2.1 Media
Nutrient Broth (NB) was made by dissolving 10 g
peptone (Beijing Oboxing Biotechnology Co., Ltd,
China), 3 g beef extract (Beijing Oboxing
Biotechnology Co., Ltd, China), and 5 g sodium
chloride in 1 L distilled water. pH was adjusted to 7.4
with 1 mol/L sodium hydroxide solution, then
sterilized at 121
o
C for 15 min. Nutrient Agar (NA)
consisted of NB with the addition of 1.5%-1.7% agar.
2.2 Samples and Sensory
Observation
Beijing sausages produced on the same day were
obtained from the Pengcheng food branch of
BeijingShunxin Agricultural Limited Company, with
the specification of weighing 350-400 g/piece. Ten
Beijing sausages were put into an open homogeneous
sack and stored at 4
o
C. The sensory indexes of the
samples were observed and recorded in time during
storage. According to the national standard SB/T
10481-2008 (Ministry of Commerce of the People's
Republic of China, 2009), a sensory record table was
set up. The results are shown in Table 1. After sensory
evaluation, they were taken out for microbiological
analysis.
Table 1: Sensory changes of Beijing sausages in storage.
Storage days/d
1-3 4 5 6 7 8 9 10 11 12 13 14 15
Color ++++ ++++ +++ +++ ++ ++ ++ ++ + + +
—
—
Texture ++++ +++ +++ ++ ++ ++ ++ + + + +
—
—
Smell ++++ ++++ +++ +++ +++ ++ ++ ++ + + + + —
(Notes: ++++: very bright color, tight and resilient tissue, no mucus and mildew, no odor and rancidity, with meat flavor;
+++: bright color, tight tissue, no mucus, and mildew, with meat flavor, no odor and rancidity; ++: dark color, whitened
epidermis, soft tissue, no meat flavor; +: dark color, whitened epidermis, soft and inelastic tissue, mild rancidity odor; —:
with white odor, juicy phenomenon, and rancid flavor.)
2.3 Analysis of the Dominant
Spoilage Bacteria in Beijing
Sausages
2.3.1 Sample Pretreatment
As shown in Table 1, when stored at 4
o
C, the samples
had reached complete spoilage after 15 days. The
spoiled sausages were evenly divided into two
groups. Under sterile conditions, each sausage’s
epidermis and central part were evenly sampled with
sterile scissors. The samples from each group were
cut and mixed thoroughly.
2.3.2 Microbiological Culture
From each of the two mixed samples, 25 g was moved
into the aseptic homogenization bag with filter
membrane, 225 mL 0.85% sterile physiological saline
solution added, and samples homogenized for 150 s
by the beat nomogenizer. Next, 1 mL of filtrate was
extracted with a pipettor, slowly injected into a test
tube containing 9 mL sterile physiological saline
solution along the tube wall, and shaken evenly on the
oscillator to make 10
-2
samples of homogenization
solution. Finally, the required concentration was
diluted 10 times in a turn, according to the estimation
of sample contamination, 2-3 samples with the
appropriate dilution were selected, and 100 μL of
sample homogenate was injected into the surface of
the medium by pipettor. Samples were allowed to set
for 10 min then incubated at 36±1
o
C for 48 h.
2.3.3 Isolation, Purification, and
Morphological Analysis of Dominant
Spoilage Bacteria
Typical single colonies cultured on NA medium were
selected and repeatedly streaked on the medium for
separation and purification to obtain pure colonies.
For the four isolated and purified individual colonies,
colony morphology was observed and recorded,
including shape, size, color, luster, transparency, edge
shape, surface smoothness, wettability, and uplift
degree. Single colonies were stained with safranin,
and the morphology of the cells was observed by
ZEISS Axioplan 2 optical microscope (10 times
eyepiece × 100 times objective lens).
Analysis of Dominant Spoilage Bacteria in Beijing Sausages
85

2.3.4 Identification of Dominant Spoilage
Bacteria by 16S rDNA Sequence
Analysis
The four strains purified as described above
(numbered R1, R2, R3, R4) were inoculated in 5 mL
corresponding medium and cultured at the
appropriate temperature.
(1) Genomic DNA extraction: Briefly, bacterial
solution (1.0 mL) was centrifuged at 13000 rpm for 2
min in a 1.5 mL centrifuge tube. The supernatant was
discarded, the pellet transferred to a fresh tube, and
1.0 mL 0.85% NaCl was added. The tubes were then
centrifuged at 13000 rpm for 2 min. The precipitate
was suspended in 550 μL 1×TE. Then, 17 μL
lysozyme (35 mg/mL), 3 μL proteinase K (20
mg/mL), and 30 μL 10% SDS were added, with 30
min incubation at 37
o
C after each. 100 μL NaCl (5
mol/L) and 80 μL CTAB/NaCl solution were added
and mixed well, and samples incubated at 65
o
C in a
water bath for 10 min. An equal volume of
chloroform: isoamyl alcohol (24:1) was added and
samples mixed. After mixing, the tubes were
centrifuged at 13000 rpm for 10 min at room
temperature. The aqueous layer was transferred to a
fresh tube, and 2 volumes of cold ethanol were added.
The tubes were centrifuged at 13000 rpm for 10 min.
The supernatant was discarded and placed at room
temperature for 30 min to remove residual ethanol.
DNA was dissolved in sterile water or TE.
(2) PCR amplification: Bacterial DNA extracted
as described above was used as the template. PCR
was performed with 16S rDNA universal primers
(forward primer 27F: 5'-AGA GTT TGA TCC TGG
CTC AG-3', reverse primer 1541R: 5'-AAG GAG
GTG ATC CAG CC-3').
Reaction conditions for PCR were as follows: 95
oC for 5 min; 30 cycles of 1 min at 95
o
C, 1 min at 57
o
C, and 1 min 20 s at 72
o
C; 72
o
C for 5 min.
The reaction mixture was 100 μL and consisted of
0.8 μL Taq (5 U/μL), 10 μL 10× PCR Buffer (Mg
2
+
Plus), 8 μL dNTP Mixture (2.5 mM/each), 2.5 ng
template DNA, 2 μL each forward and reverse
primers (10 μmol/L), and ddH
2
O to 100 μL (ARa et
al., 2006; Cheng et al., 2006).
(3) Sequence alignment: The PCR amplification
products were purified and sequenced by Beijing
Haocheng Mingtai Technology Co., Ltd. The 16S
rDNA sequence of the strains with high homology
were downloaded from NCBI and analyzed by
MEGA 7 software.
(4) Phylogenetic analysis: Phylogenetic analysis
was performed by MEGA 7.0.26 software. The
phylogenetic tree was built using the neighbor-
joining method and with 1000 bootstrapping
replicates.
2.4 PCR-DGGE Analysis of Spoilage
Bacteria in Beijing Sausages
Bacteria were identified using PCR denaturing
gradient gel electrophoresis. Twenty-five grams of
the pretreated sample described in Section 2.3.1 was
packed in a sterile homogenization sack, added 225
mL 0.85% sterile physiological saline solution, and
the sample homogenized for 150 s. Approximately 4
mL of sample was transferred to a sterile 5 mL
centrifuge tube. After storage at -80 oC for 24 h,
samples were sent to the Beijing Haocheng Mingtai
Technology Co., Ltd for PCR-DGGE identification.
The DGGE recovered bands were PCR amplified.
3 RESULTS
3.1 Identification Results and Analysis
of 16S rDNA
Morphological characteristics of strains R1-R4 are
shown in Table 2. The microscopic images of the
safranin single dye are shown in Figure 1.
Table 2: Morphology characteristics of different strains.
Colonial morphology R1 R2 R3 R4
Shape circle circle circle circle
Size big small small small
Color light milky white white white white
Luster glossy glossy glossy dim
Transparency translucent opaque opaque opaque
Edge shape regular regular regular regular
Sshirunshiruurface smooth smooth smooth smooth
Wettability moist dry moist moist
Uplift raised flat raised raised
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
86
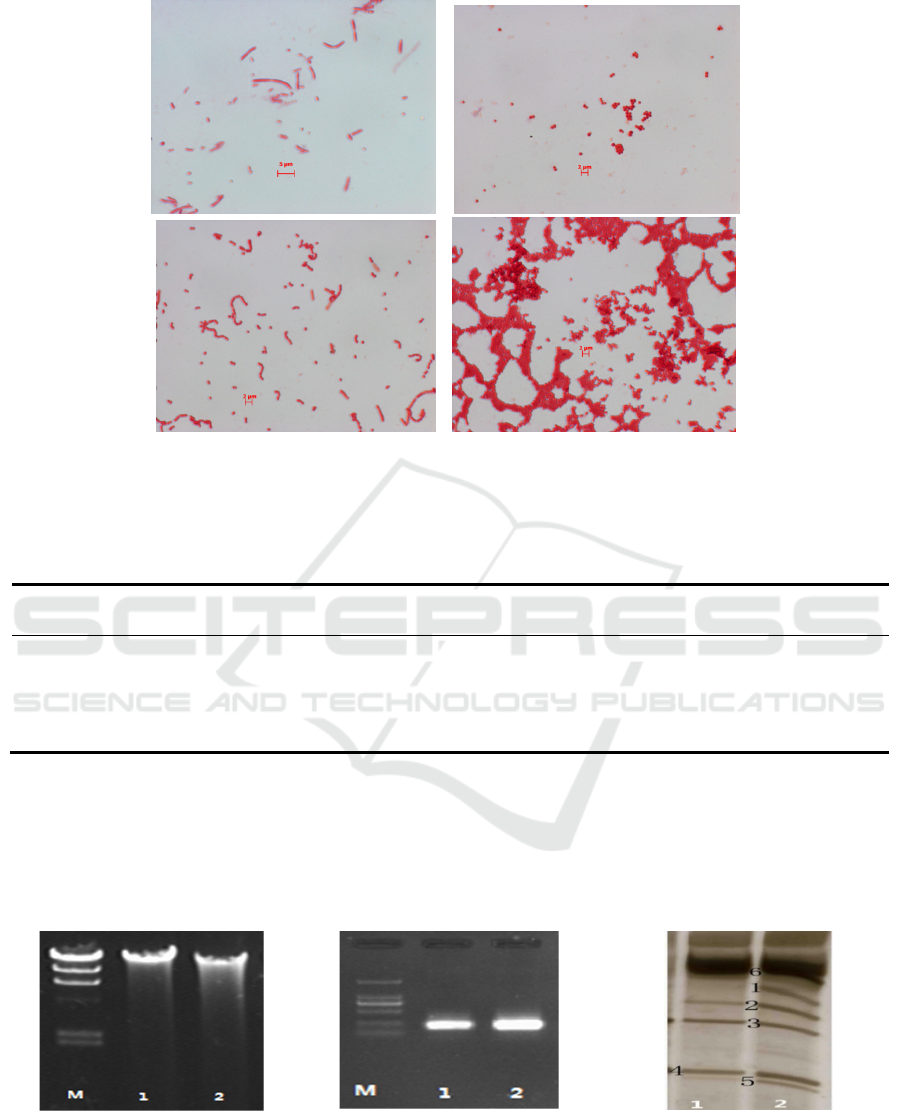
a b
c d
Figure 1: Microscope images of strains R1 (a), R2 (b), R3 (c) and R4 (d).
Four strains were isolated and purified for 16S
rDNA sequence analysis. NCBI was used to identify
the bacteria based on the sequences; analysis results
are shown in Table 3.
Table 3: Result of NCBI sequence comparison analysis of four strains.
. Strain label Similarity
Population and login number of the strain
with the highest similarity in GenBank
English name
1 R1 99.78% Pseudomonas lundensis ATCC 49968(T) Pseudomonas sp.
2 R2 100% Staphylococcus vitulinus ATCC51145(T) Staphylococcus sp.
3 R3 100% Brochothrix thermosphacta ATCC 11509(T) Brochothrix sp.
4 R4 100% Brochothrix thermosphacta ATCC 11509(T) Brochothrix sp.
3.2 Identification
Results and
Analysis of PCR-DGGE
The result of DNA extraction is shown in Figure 3.
The result of PCR amplification is shown in Figure 4.
DGGE patterns of bacterial DNA are shown in Figure
5. The primer was 16S rDNA corresponding to the
universal primer 338F-534R, and the PCR product in
the map was 250bp, which was judged as the target
fragment. The PCR amplification results are shown in
Figure 4.
Figure. 3: Result of DNA extraction. Figure. 4: Result of PCR amplification. Figure. 5: DGGE patterns of bacterial
DNA.
(Notes: The Arabic numerals in the figure indicate the number of the rubber recovery, with only one band cut at the same
level. Due to the low brightness of band 6 in DGGE, the dominant bacteria were not considered, and the gray value analysis
was not carried out.)
Analysis of Dominant Spoilage Bacteria in Beijing Sausages
87

4 CONCLUSIONS
Two methods, microbial cultivation and polymerase
chain reaction-denaturing gradient gel
electrophoresis (PCR-DGGE), were used to analyze
the dominant spoilage bacteria in Beijing sausages.
Using 16S rDNA sequence analysis, the main
spoilage bacteria isolated and identified were one
genus of Pseudomonas sp., one genus of
Staphylococcus sp., and two genera of Brochothrix
sp. PCR-DGGE analysis identified 13 species of
bacteria, including Pseudomonas sp., Brochothrix sp.,
and Staphylococcus sp., with Pseudomonas sp. being
the most dominant spoilage bacteria, followed by
Brochothrix sp. and Staphylococcus. The results of
the two methods are consistent with one another and
with the results of other studies in the past two years
identifying spoilage bacteria in low-temperature meat
products (Samelis et al., 2000; Jenni et al., 2015).
ACKNOWLEDGMENTS
This work was supported by Beijing Key Laboratory
of Agricultural, Product Detection and Control of
Spoilage Organisms and Pesticide. The authors
express their thanks to Beijing Innovation
Consortium of Swine Research System and
Pengcheng food branch of Beijing Shunxin
Agricultural Limited Company for providing the
primary sample.
REFERENCES
Adams M R, Moss M. Food Microbiology[M]. RSC, 2007.
ARa C, SeungKu Y, EuiJoong K. Cloning, sequencing and
expression in Escherichia coli of a thermophilic lipase
from Bacillus thermoleovorans ID‐1[J]. FEMS
Microbiology Letters, 2006,186(2).
Cheng H, Jiang N. Extremely Rapid Extraction of DNA
from Bacteria and Yeasts[J]. Biotechnology Letters,
2006,28(1).
Danilo E, Federica R, Antonella N, et al. Mesophilic and
psychrotrophic bacteria from meat and their spoilage
potential in vitro and in beef.[J]. Applied and
environmental microbiology, 2009,75(7).
Doulgeraki A I, Ercolini D, Villani F, et al. Spoilage
microbiota associated to the storage of raw meat in
different conditions[J]. International Journal of Food
Microbiology, 2012,157(2).
Jenni H, Riitta R, Javeria A, et al. Meat Processing Plant
Microbiome and Contamination Patterns of Cold-
Tolerant Bacteria Causing Food Safety and Spoilage
Risks in the Manufacture of Vacuum-Packaged Cooked
Sausages[J]. Applied and Environmental
Microbiology, 2015,81(20).
Liu F, Wang D, Du L, et al. Diversity of the Predominant
Spoilage Bacteria in Water‐Boiled Salted Duck during
Storage[J]. Journal of Food Science, 2010,75(5).
Ministry of Commerce of the People's Republic of China.
SB/T 10581-2008, Quality safety requirement of
pasteurized meat products[S]. Beijing : Standards Press
of China, 2009.
Peirson M D, Guan T Y, Holley R A. Aerococci and
carnobacteria cause discoloration in cooked cured
bologna [J]. Food Microbiology, 2003,20(2).
Samelis J, Kakouri A, Rementzis J. Selective effect of the
product type and the packaging conditions on the
species of lactic acid bacteria dominating the spoilage
microbial association of cooked meats at 4°C[J]. Food
Microbiology, 2000,17(3).
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
88
