
Research on the Impact of Alkylbenzene Sulfonate Surfactants in Use
in Oilfields on Native Biodiversity of Mining Areas and Response to
Environmental Remediation of Native Biodiversity
Zhanyou He
1,2,a,*
, Ziqian Xu
1,2,b
, Min Zhao
1,2,c
, Yan Li
1,2,d
and Ning Liu
1,2,e
1
Research Institute of Oil and Gas Technology of Changqing Oilfield Company, China
2
National Engineering Laboratory for Exploration and Development of Low-permeability Oil and Gas Field, China
d
liyan001_cq@petrochina.com.cn,
e
liun15_cq@petrochina.com.cn
Keywords:
Alkylbenzene Sulfonate, Surfactants, Soil Bacteria, High-Throughput Sequencing.
Abstract:
In order to clarify the effect of alkylbenzene sulfonate surfactants used in oil field on native biodiversity in
mining area. In this paper, high-throughput sequencing and biodiversity analysis were performed on the soil
continuously polluted by alkylbenzene sulfonate surfactants. Clostridia, BPC102 and Bacteroidia became the
dominant bacteria in the soil environment, with strong self-repair response of environmental organisms.
Bacteria S035, Bacilli and Dothideomycetes showed negative response, indicating that alkylbenzo sulfonate
surfactants inhibited and affected the growth, development and reproduction of these native organisms. The
results showed that alkylbenzene sulfonate surfactant had obvious effects on soil biodiversity in mining area,
which provided scientific basis for environmental impact assessment and environmental management of
surfactants.
1 INTRODUCTION
Alkylbenzene sulfonate surfactants are widely used
in the field of chemical flooding in low-permeability
oilfields for enhanced oil recovery due to their
excellent oil displacement performance. As an
important oil field EOR chemical agent, the
application range of surfactants is still expanding and
the consumption is also increasing day by day. In the
process of use, a large amount of waste water and
waste residues containing surfactants are inevitably
discharged and infiltrated into the soil. Large-scale
industrial use of alkylbenzene sulfonate surfactants
urgently needs to clarify its impact on the native
biodiversity of oilfields and mining areas and the
response of native biodiversity to its environmental
bioremediation.
The environmental behavior of alkylbenzene
sulfonate surfactants in soil mainly includes
migration, adsorption and degradation. As an
important place for energy exchange of various
substances recycling machines, soil is usually the
destination of migration, retention and deposition of
pollutants in the environment. After surfactants enter
the mining environment, they will first have a certain
impact on the biodiversity of the mining area. The
impact degree is positively correlated with the impact
of local biodiversity, and negatively correlated with
the bioremediation response of local biodiversity. If
the native biodiversity has a strong response to the
environmental bioremediation of alkylbenzene
sulfonate surfactants, it indicates that the oil field has
a high tolerance of alkylbenzene sulfonate surfactants
and a large marginal safety concentration, that is, the
environmental biological reference value is large, and
the environmental biological toxicity is small. On the
other hand, if the native biodiversity responds weakly
to the environmental bioremediation of alkylbenzene
sulfonate surfactants. The microbial flora that can
degrade the surfactant cannot be enriched in a short
period of time, the tolerance of alkylbenzene
sulfonate surfactants in the oilfield will be low and
the safety marginal concentration will be small. In
other words, alkylbenzene sulfonate surfactants have
low environmental biological reference value and
high environmental biological toxicity in oilfield
mining area. Therefore, it is of great significance to
study the effects of alkylbenzene sulfonate
surfactants on native biodiversity and the response of
native biodiversity to environmental bioremediation.
He, Z., Xu, Z., Zhao, M., Li, Y. and Liu, N.
Research on the Impact of Alkylbenzene Sulfonate Surfactants in Use in Oilfields on Native Biodiversity of Mining Areas and Response to Environmental Remediation of Native Biodiversity.
DOI: 10.5220/0011211200003443
In Proceedings of the 4th Inter national Conference on Biomedical Engineering and Bioinformatics (ICBEB 2022), pages 379-384
ISBN: 978-989-758-595-1
Copyright
c
2022 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
379

In this paper, different types of bioreactors with
alkylbenzene sulfonate surfactants as the only
pollution source have been designed and
continuously operated. The second-generation
Qualcomm sequencing technology (Illumina MiSeq)
was used to conduct high-throughput sequencing of
16S rDNA V3~V4 regions and ITS1 regions on the
soil which was continuously polluted by
alkylbenzene sulfonate surfactants. The sequencing
results were evaluated by OTU cluster analysis,
Alpha diversity, species composition and abundance
analysis methods, which provided a theoretical basis
for the ecological and environmental protection in the
oilfields and mining areas.
2 METERIALS AND METHODS
2.1 Experimental Materials
Experimental target material: alkylbenzene sulfonate
surfactant used in an oilfield
Experimental soil: Fresh soil randomly collected
from a domestic oilfield chemical flooding enhanced
oil recovery test mining area, remove surface rocks
and other impurities, mix well and pass through a
2mm sieve.
Experimental equipment: In order to obtain the
influence of alkylbenzene sulfonate surfactants on
local biodiversity after entering the soil environment
and the response of local biodiversity to
bioremediation of characteristic pollution sources, a
bioreactor was designed as shown in Figure.1. The
prepared surfactant solution is continuously
introduced into the reactor soil from the top of the
bioreactor. The bottom is provided with an outlet
from which the solution can seep out. The
surrounding sampling holes are used to collect soil
samples at different contamination times.
Figure 1: Schematic diagram of bioreactor.
2.2 Experimental Methods
A peristaltic pump was used to add the prepared
surfactant solution at a certain flow rate (calculated
based on the actual leakage) from the top of the
reactor, and keep the experimental temperature
relatively constant. Samples were taken on day 7
(represented by D), day 30 (represented by E) and day
60 (represented by F), and set a group of blank
samples for control (represented by V) at the same
time. The obtained soil samples were stored in
sterilized sealed bags at −80 °C and microbial
sequencing was performed as soon as possible. The
whole experiment was carried out under dark
conditions.
3 RESULTS AND ANALYSIS
3.1 OTU Cluster Analysis
In order to facilitate analysis in the study, a single
marker is artificially set for a Taxonomic unit, namely
OTU (Operational Taxonomic Units). In order to
understand the number of species and genera in the
sequencing results of a sample, it is necessary to
classify the sequence. Through the classification
operation, sequences are divided into many groups
according to their similarity, and one group is an
OTU.
Figure.2 is a Venn diagram of the number of
bacterial OTU in soil sample. As shown in the figure,
a total of 5276 OTU were obtained in group D, 4951
OTU in group E, 5078 OTU in group F, and 4600
OTU in group V of control group. The sequence of
OTU numbers in the four soil samples is D > F > E >
V. The richness of bacterial groups was the highest in
the day7, and the lowest in the blank group. The
number of OTUs in the four groups was compared in
pairs: there were 2694 OTUs shared by group V and
Group D, 2413 OTUs shared by group V and Group
E, 2204 OTUs shared by group V and group F, 2815
OTUs shared by group D and group E and 2573
OTUs shared by group E and group F. It can be seen
that the bacterial groups in the soil samples of day7
and day30 had the highest consistency and the
smallest difference, while day60 had the lowest
consistency and the largest difference. This indicated
that as the pollution time of the alkylbenzene
sulfonate on the soil is prolonged, the difference of
the bacterial groups in the soil is greater.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
380
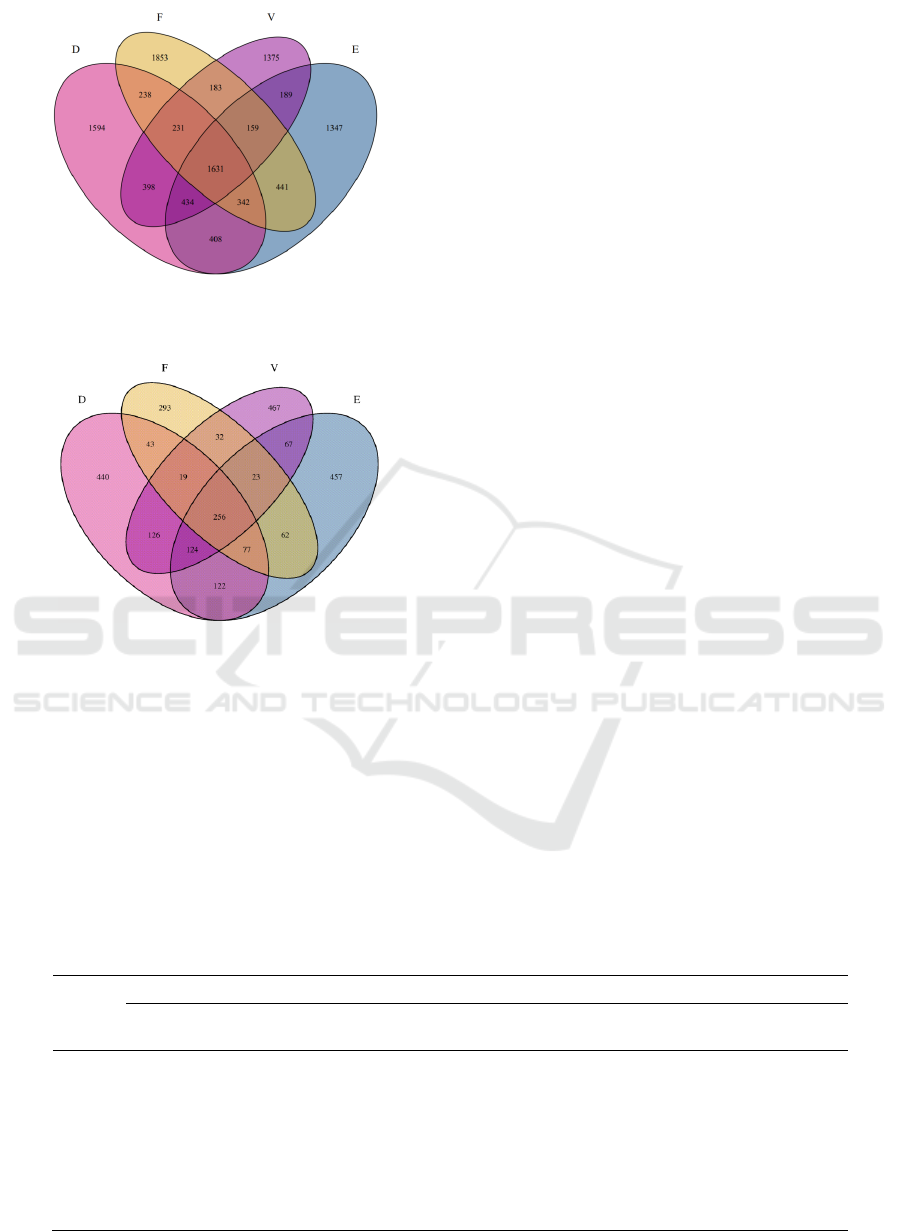
Figure 2: The Venn diagram of the number of bacterial
OTUs in soil samples.
Figure 3: The Venn diagram of the number of fungal OTUs
in soil samples.
As shown in the figure, there were 1114 OTUs in
the control group, 1207 OTUs in the D group, 1188
OTUs in the E group and 805 OTUs in the F group.
The number of OTU in the four groups of soil
samples was D > E >V > F. Fungal species richness
was highest in soil samples after 7 days of
contamination, and lowest in soil samples after 60
days of contamination. The pairwise comparison
results show that the number of OTUs shared by
group V and D is 525, the number of OTUs shared by
group D and E is 579, the number of OTUs shared by
group E and F is 418, and the number of OTUs shared
by group V and F is 418. The total number of OTUs
is 470, the number of OTUs shared by groups V and
F is 330, and the number of OTUs shared by the 4
groups of soil samples is 256. After comparison, it
was found that the consistency of fine fungal groups
was the highest and the difference was small between
the soil samples at day7 and day30. While the
consistency was the lowest and the difference was the
largest between the soil samples at day60 and the
control group. It can be seen that with the extension
of time, the difference of fungal groups in soil
contaminated by alkylbenzene sulfonate surfactant
gradually increased.
3.2 Alpha Diversity Index Analysis
MiSeq platform was used to perform high-throughput
sequencing on four groups of soil samples by
sequencing while synthesizing. The sequencing
results are shown in Table1. Shannon index and
Simpson index are usually used to estimate the
diversity of OTU species in microbial communities.
They are also commonly used to estimate the
diversity of microorganisms in sample. The larger the
Shannon index value, the higher the community
diversity. The smaller the Simpson index value, the
higher the community diversity. It can be seen from
Table1 that the order of the species diversity of
bacteria and fungi in the four groups of soil samples
is V> D> E> F. This shows that as time goes by, the
microbial diversity of the soil contaminated by
surfactants has declined, and the surfactants have an
inhibitory effect on the growth of microorganisms.
The coverage of each sample was more than 99.90%,
indicating that the sequencing effect was ideal, and
the diversity analysis results fully reflected the
information of micro
bial species in the area.
Table 1: Diversity analysis of bacterial microbial index of soil samples.
Group
Bacteria Fungi
Shannon Simpson
Coverage,
%
Shannon Simpson
Coverage
,%
D
7.12±
0.115
0.0018±
0.00035
99.91 4.82 ±0.285 0.0236 ±0.00890 99.95
E
6.90±
0.393
0.0042±
0.00483
99.93 4.52 ±0.586 0.0410 ±0.04184 99.94
F
6.71±
0.904
0.0093±
0.01571
99.92 3.68 ±1.320 0.1398 ±0.17308 99.96
V
7.18±
0.085
0.0014±
0.00018
99.91 4.96 ±0.193 0.0180 ±0.00412 99.94
Research on the Impact of Alkylbenzene Sulfonate Surfactants in Use in Oilfields on Native Biodiversity of Mining Areas and Response to
Environmental Remediation of Native Biodiversity
381
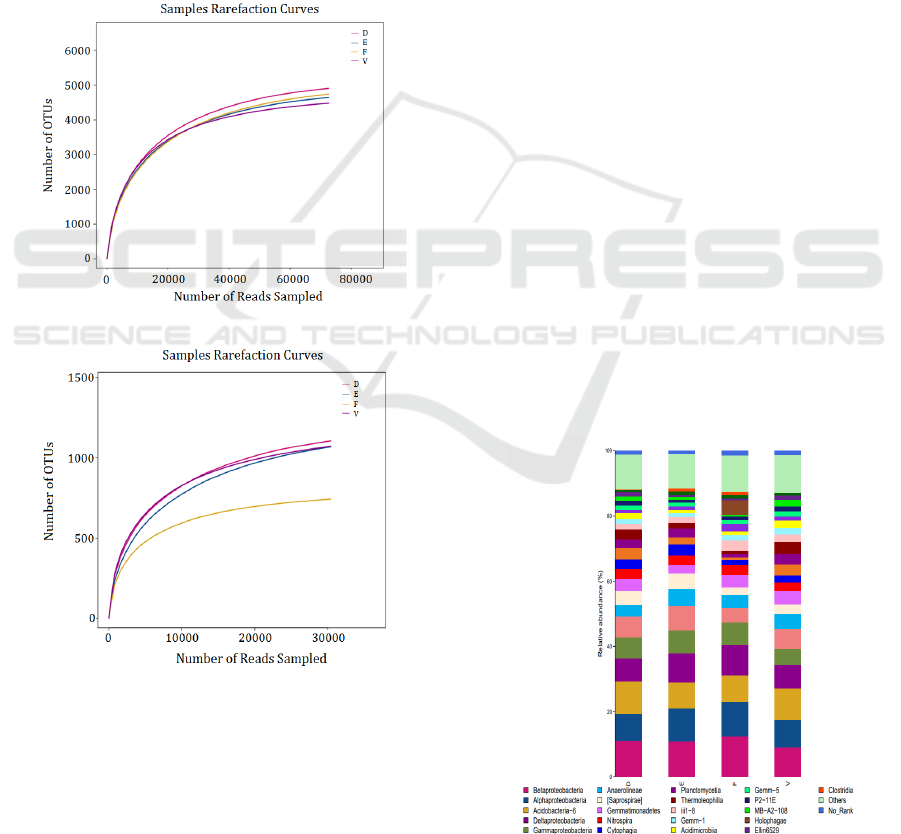
Random sampling of sequencing sequences is
used to construct a curve based on the number of
extracted sequences and the number of OTU
represented by them, that is the dilution curve.
Generally, when the curve tends to be flat, it indicates
that the number of samples is reasonable.
Figure.4(a)and Figure.4(b) are the dilution curves of
soil samples. It can be seen that the dilution curves of
bacteria and fungi of the samples are basically flat,
indicating that the sequencing and sampling are
reasonable and can truly reflect the microorganisms
in the soil samples. Combined with the coverage of
each sample, it shows that most of the microbial
groups are included in the sequencing results, which
can truly reflect the composition of the microbial
community in the soil in this area.
(a) Bacteria
(b) Fungi
Figure 4: Dilution curve of the sample.
3.3 Analysis of Community Structure
As shown in Figure.5, more than 70 types of bacteria
were detected in the four groups of soil samples.
Among them, β-Proteobacteria, α-Proteobacteria,
Acidobacteria, Δ-Proteobacteria and γ-Proteobacteria
are the absolute dominant species. The proportions of
the five dominant bacteria in the blank group were
9.04%, 8.55%, 9.62%, 7.16%, and 4.96%,
respectively. By day 7, the proportions of these five
bacteria were 11.04%, 8.35%, 9.97%, 7.01% and
6.41%. By day 30, the proportions of the five
dominant bacteria were 10.95%, 10.14%, 7.90%,
8.93%, and 7.13%. By day 60, the proportions of the
five bacteria were 12.46%, 10.50%, 8.24%, 9.24%
and 6.92%, respectively. In general, the relative
content of the five dominant bacteria did not change
much with the prolongation of pollution time, and
their growth and reproduction in the soil were
relatively stable, and they were not greatly affected
by alkylbenzene sulfonate surfactants.
Figure.6 is a Heatmap based on the genus level.
The Heatmap can reflect the similarity and difference
in species composition of all samples at a specific
taxonomic level. The more similar species or samples
are, the closer they are to each other in the cluster tree,
which can also indicate that certain bacterial groups
may have specific distributions.
The relative abundance of Clostridia in soil
samples at day30 and day60 was higher than day7 and
black group. Similarly, the relative abundances of
bacteria BPC102 and Bacteroidia were higher in the
soil samples on day60 than in other soil samples,
indicating that under the continuous pollution of
alkylbenzene sulfonate, Clostridia, BPC102 and
Bacteroidia gradually became the dominant bacteria
in the soil. However, the relative abundance of
bacteria S035 and Bacilli decreases with the
extension of the experiment time, indicating that the
growth and propagation of bacteria S035 and Bacilli
are inhibited by alkylbenzene sulfonate.
Figure 5: Histogram of bacterial community composition at
class classification level.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
382

Figure 6: Heatmap at class classification level.
Figure 7 is a histogram of fungal community
composition based on class classification level. More
than 20 types of fungi were detected in the four
groups. Among them, the relative proportions of
Sordariomycetes, Mortierellomycetes and
Agaricomycetes were 21.49%~28.03%,
10.81%~27.95%, 13.29~19.62%, which were
relatively high in soil samples, and were the absolute
dominant fungal species. The relative content of
Dothideomycetes in the blank group was 10.15%,
which decreased to 2.02% on day 60, indicating that
alkylbenzene sulfonate would inhibit the growth and
reproduction of Dothideomycetes.
Figure 7: Histogram of bacterial community composition at
class classification level.
Figure 8: Heatmap at class classification level.
4 CONCLUSION
In this paper, the oil field soil continuously polluted
by alkylbenzene sulfonate surfactants was taken as
the research object. The results of high-throughput
sequencing and biodiversity analysis were as follows:
(1) As the soil is continuously polluted by
alkylbenzene sulfonate surfactants for a longer time,
the abundance and diversity of bacteria and fungi in
the soil are decreasing, and the group differences of
microorganisms are gradually increasing. It shows
that alkylbenzene sulfonate surfactants have a
significant impact on the native biodiversity in the
soil environment of mining areas.
(2) Microbial sequencing showed that
alkylbenzene sulfonate contaminated soil had
dominant bacteria, They were Betaproteobacteria,
Alphaproteobacteria, Alphaproteobacteria,
Acidobacteria, Deltaproteobacteria,
Gammaproteobacteria, Clostridia, BPC102 and
Bacteroidia.
(3) Clostridia, BPC102 and Bacteroidia had
positive response to alkylbenzene sulfonates, and
gradually became the dominant bacteria in the soil
environment of the oil field. It showed that the native
biodiversity of the oilfield mining area had a strong
environmental biological self-repair response to
alkylbenzene sulfonate surfactants.
(4) The relative content of bacteria S035, Bacilli,
and fungus Dothideomycetes (Polycystomycetes)
decreased with the extension of the contamination
time, that is, it showed a negative response to alkyl
Research on the Impact of Alkylbenzene Sulfonate Surfactants in Use in Oilfields on Native Biodiversity of Mining Areas and Response to
Environmental Remediation of Native Biodiversity
383

benzene sulfonates. The results showed that
alkylbenzenesulfonate surfactants inhibited and
affected the growth, development and reproduction of
these native organisms.
It can be seen that alkylbenzene sulfonate
surfactant have an obvious impact on the diversity of
soil biodiversity in mining areas. Therefore, their use
and discharge must consider environmental capacity,
fundamentally reduce their direct discharge to the
environment, and increase the treatment of
wastewater and waste residue containing surfactants.
REFERENCES
Cao HJ, Ni HW (2015). Soil microbial diversity and its
influencing factors: A review [J]. Land and Natural
Resources Research, (03):85-88.
He Z, Zhao TT, Xing ZL, Yuan JH (2015). Microbial
Community analysis of typical domestic waste landfill
cover soil [J]. China Environmental Science. Vol. 35
(12): 3744-3753
Jia H B (2013). Microbial remediation of petroleum
contaminated soil and its effects on soil bacterial
community diversity [D]. Northeast Forestry
University.
Jiang SY (2016). Research on microbial diversity in major
oil shale mining areas in my country [D]. Northeastern
University.
Kuang L, Zhang X.J, Wang BH, Ji Wi, Sui X, Kan LB
(2008). Environmental behavior and harm analysis of
surfactants in soil [J]. Journal of Northeast Forestry
University, VOI.36 No. 2
Niu SQ, Long Y, Li HY, Da WY, Hu S, Li WJ, Zhu XT,
Kong WB (2017). IlluminaMiSeq high-throughput
sequencing technology for analysis of microbial
diversity in saline soil of Hexi Corridor [J]. Chinese
journal of microbiology ,44(09):2067-2078.
Qin N, Li DF, Yang RF (2011). High throughput sequencing
technology and its application in microbiology [J]. Acta
microbiologica sinica, 51(04): 445-457.
ICBEB 2022 - The International Conference on Biomedical Engineering and Bioinformatics
384
