
AN USER-CENTRIC AND SEMANTIC-DRIVEN QUERY
REWRITING OVER PROTEOMICS XML SOURCES
Kunalè Kudagba, Omar El Beqqali
MISI Research Group, USMBA University, Dhar El Mehraz Faculty of sciences, Fes, Morocco
Hassan Badir
SIC Department, National School of Applied Science (ENSA), Tangier, Morocco
Keywords: Proteomics, Ontology, XML, Trees, Semantic Web, Query Rewriting, Minimal Transversals.
Abstract: Querying and sharing Web proteomics data is not an easy task. Given that, several data sources can be used
to answer the same sub-goals in the Global query, it is obvious that we can have many candidates
rewritings. The user-query is formulated using Concepts and Properties related to Proteomics research
(Domain Ontology). Semantic mappings describe the contents of underlying sources. In this paper, we
propose a characterization of query rewriting problem using semantic mappings as an associated
hypergraph. Hence, the generation of candidates’ rewritings can be formulated as the discovery of minimal
Transversals of an hypergraph. We exploit and adapt algorithms available in Hypergraph Theory to find all
candidates rewritings from a query answering problem. Then, in future work, some relevant criteria could
help to determine optimal and qualitative rewritings, according to user needs, and sources performances.
1 INTRODUCTION
The rapid progress of biotechnologies and the
multiple genome project (Davidson, 1995) (Bishop,
1999) about organisms and diverse species have
generated an increasing amount of proteomic data
stored in many sources (Kohler, 2004), available and
publicly accessible on the Web. They contain data
about metabolic pathways, protein 3D structures,
DNA Sequences, organisms, diseases, and so on.
Many biological questions require that data from
several data sources are queried, searched, and
integrated. The first step in this process of Data
Integration is the Query rewriting which consists of
reformulating a global query in several specific local
queries. The query rewriting problem has recently
received significant attention because of its
relevance to a wide variety of data management
problems (Halevy, 2001): query optimization,
maintenance of physical data independence, data
integration, and data warehouse design.
Semantic mappings can be used to adapt a
global query expressed in terms of a Domain
Ontology in terms of specific local sources. In fact,
as several sources could provide expected resources,
more than one could be relevant to rewrite the global
query.
The motivation of this work is to investigate how
to generate candidate rewritings when user-query is
posed over XML Sources, and mappings are
expressed in LAV Approach. To achieve this goal,
we characterize the query rewriting problem as
minimal Transversals Discovery from an associated
Hypergraph. From this set of generated rewritings,
we would compute an optimal and best quality
rewriting based on some defined and relevant
criteria. We illustrate an intuitive execution of the
rewriting algorithm proposed, using a scenario of
Proteomics data sources.
The following two XML sources contain data
that are semantically similar, but are described with
autonomous and heterogeneous schemas. They both
represent the same proteomics data, but not
identically. They give an idea of differences in terms
of terminologies, structures and contents.
123
Kudagba K., El Beqqali O. and Badir H. (2009).
AN USER-CENTRIC AND SEMANTIC-DRIVEN QUERY REWRITING OVER PROTEOMICS XML SOURCES .
In Proceedings of the 11th International Conference on Enterprise Information Systems - Databases and Information Systems Integration, pages
123-130
DOI: 10.5220/0001986301230130
Copyright
c
SciTePress

XML Data Source 1
XML Data Source 2
We can remark also some semantics
heterogeneities, like ACC_NUMBER attribute in
Source 2 which is equivalent with ACCESSION
Element in Source 1.
Although both data sources contain semantically
similar proteomic data, the simple user query
”Which are proteins that are encoded by the gene
named by CSF2RB2 in Mus Musculus Organism? ”
need to be formulated quite differently with existing
XML query languages, like Xquery or XPath for
both sources.
So, flexible, user-centric and semantic
strategies of discovering relevant sources are needed
to compute optimal and best quality rewriting,
according to suitable criteria.
The rest of the paper is structured as follows.
Section 2 gives a brief survey on our related work
regarding existing query rewriting approach in
Mediators. Section 3 discusses the basic concept of
Knowledge representation. The hypergraph-based
semantic query rewriting algorithm is presented in
Section 4. The last section 5 draws the conclusions
and future work.
2 RELATED WORK
One of the first LAV systems that allow the
integration of XML is AGORA (Manolescu, 2001).
But AGORA still makes an extended use of the
relational model: Although it offers an XML view
for relational and XML data, this view is translated
into a generic relational schema, XML resources are
described as relational views over this schema and
XQuery expressions are translated to standard SQL
queries, which are then decomposed and evaluated.
Information Manifold (Levy, 1996) also follows
a local-as views approach. In this system, the global
schema is a flat relational schema, and Description
Logics are used to represent hierarchies of classes.
The sources are expressed as relational views over
this schema. Query rewriting is done by the Bucket
algorithm which rewrites a conjunctive query
expressed of the global schema using the source
views. It examines independently each of the query
sub-goals and tries to find rewritings but loses some
by considering the sub-goals in isolation.
STyX system (Fundulaki, 2002) already uses a
domain Ontology as global schema language and
translates an OQL-like global query language to
XQuery expressions on the heterogeneous XML
sources. STyX maps XPath expressions to ontology
concepts. (Amann, 2002) discuss a data integration
system whereby XML sources are mapped into a
simple ontology (supporting inheritance and roles,
but no description logic-style definitions).
In (Lehti, 2004) authors have also used to
integrate XML heterogeneous data sources. Their
work consists to map XML schema constructs to
concepts. The main difference to STyX is the
approach to semantic mapping. Although Lehti’s
approach is not as flexible and powerful as using
XPath mappings, it is in principle able to detect
inconsistencies in the mapping with the help of a
description logic reasoner (Baader¸ 2003).
Other data integration approaches that use an
Ontology as Global Schema, are either based on an
extended data warehouse. Semantic mediation in C-
Web (Baader¸ 2003) is based on thesauri. In Xyleme
mediator (Delobel, 2003), the global schema is a set
of abstracted DTDs which are terms Trees according
to domain vocabularies such as Culture or Tourism.
Both follow GLAV (GAV and LAV together)
because correspondences between Mediator
vocabulary and Sources vocabularies are expressed
by simpler path mappings.
Recently, with the development of Semantic
Web, mediation systems have been developed.
Project Piazza (Halevy, 2003) proposes an
<PROTEIN_SET>
<PROTEIN>
<ACCESSION>P26954</ACCESSION>
<ENTRY_NAME>IL3B_MOUSE</ENTRY_NAME>
<PROTEIN_NAME>Interleukin-3 receptor
class II beta chain [Precursor]
</PROTEIN_NAME>
<GENE_NAME>CSF2RB2</GENE_NAME>
<ORGANISM taxonomy_id="10090">Mus
musculus</ORGANISM>
</PROTEIN>
…
</PROTEIN_SET>
<PROTEIN_BASE>
<PROTEIN ACC_NUMBER="P26954 >
<ENTRY>IL3B_MOUSE</ENTRY>
<PROTEIN_NAME>Interleukin-3 receptor
class II beta chain [Precursor]
</PROTEIN_NAME>
<GENE>CSF2RB2</GENE>
<ORGANISM >
<TAX_ID >10090</TAX_ID >
<NAME> Mus musculus</NAME>
</ORGANISM>
</PROTEIN>
….
</PROTEIN_BASE>
ICEIS 2009 - International Conference on Enterprise Information Systems
124

infrastructure based on Peer-to-Peer (like a
decentralized mediator) for RDF and OWL data
integration.
According to the query rewriting algorithm, you
can refer to (Halevy, 2001) for a large survey on the
Query rewriting problem. Our approach is inspired
by WS-CatalogNet’s semantic-driven Algorithm. In
this work, (Benatallah, 2006) have developed a
novel and more advanced query rewriting techniques
for flexible and effective E-Catalogs selection.
3 REPRESENTING
KNOWLEDGE
In order to rewrite semantically a global query, it is
essential to make a choice of an adequate abstraction
model for local sources and to express in a common
formalization language all available knowledge.
This last case concerns the domain ontology, the
semantic mappings, and the user query.
The proteomics sources which we are working
with are stored and available as XML Documents
according to their XML Schemas. XML (Bray,
1998) is presently becoming the standard for the
exchange of biological data sources. So, the reason
for the use of XML Sources for the data Integration
is obvious. XML Schemas (Thompson, 2000) are
more suitable than DTDs for expressing the syntax,
structural, cardinality and typing constraints required
by proteomics data. We propose to abstract sources
XML Schemas as unordered Trees, and we try to
propose a specification language based on
description logic (DL) (Baader, 2003) formalization
and reasoning (Trees Logics).
3.1 Trees Abstract Model
We know that XML Schemas are special XML
Documents. Various models have been proposed to
represent XML Documents. The W3C proposed a
generic model named Document Object Model
(DOM). In this model, presented in the current
section, XML Documents can be abstracted as
Trees. Our motivation using this way of abstraction
is to further exploit some achieved and well known
results on Trees Embedding Problem (Schielder,
2001) as knowledge semantic retrieval in an
integration framework.
3.2 Logic-based Trees Descriptions
To provide a semantic formalization, necessary for
rigorous characterisation of proteomic queries and
knowledges, we propose to use a description
language of hierarchic structures such as Trees,
based on Logics and called Trees-Logics.
Many researchers have addressed the question of
using logics over Trees. In (Deutsch, 2003 and
2005) authors have translated XQuery global queries
into local conjunctive queries over Trees in a Data
integration processes. In (Schielder, 2001) a
language called ApproXQL, which exploits among
others logical operators, to formalize more richer
and expressive requests, has been developed. These
requests expressed as conjunctive queries could be
illustrated and interpreted as Trees. Then, in the
paper (Gotlob and Koch, 2004) authors have studied
the complexity and the expressive power of
conjunctive queries over Trees.
So, we believe strongly that it is possible to
describe data Trees with a suitable subset of logical
formalisms (Baader, 2003). We want to exploit all
these works in order to provide a logical description
of hierarchical structures, such as Trees and
consequently Paths in particular. In our final
integration framework, both the phases of Trees
generation and their specification in Trees-Logics
are totally transparent for the user. We precise that
Description Logics (Baader, 2003) are a family of
logics which were developed for modeling complex
hierarchical structures and to provide a specialized
reasoning engine to do inferences on these
structures.
Due to the space limitation, we could not give
more details on Trees-Logics and so this paper will
focus only on the query rewriting problem.
4 QUERY REWRITING
In this section, we begin by presenting an abstraction
of our approach for query rewriting. Then, we show
that using some hypergraph Theory results can help
generate candidate rewritings. Therefore we present
the Classical algorithm to compute minimal
Transversals of a hypergraph. Finally, we illustrate
an execution of this alogorithm to find candidate
rewritings, given concrete case of bio-query
reformulation.
We recall that the main goal of query rewriting
phase is to reformulate a Global query
Q expressed
as Trees-Logics over Domain ontology, into Local
AN USER-CENTRIC AND SEMANTIC-DRIVEN QUERY REWRITING OVER PROTEOMICS XML SOURCES
125

queries
j
'Q
that are expressed in terms of Local
schemas. This operation is realized using semantic
mappings pre-calculated and stored on the mediator.
Semi-automatic detection of semantic mappings has
no impact on the processing time of the user query.
The concrete algorithm showing how these semantic
mappings are calculated is out of scope of this paper.
The domain Ontology is abstracted as a Tree and
expressed using the defined language, Trees-Logics.
The knowledge domain concerns proteomics
research including concepts such Protein Family, 3D
Structures, Coding Genes, Motifs, Domains, amino-
acids sequences, Active Sites, Binding Sites,
Enzymes, Chains, Chemical Bonds, … and their
relative properties, so we call the ontology by
O’proteomics. Due to space constraints, we could
not give more details on O’proteomics.
The Ontology constitutes a support for user
query formulation and gives an idea of which
concepts, it is possible (but not obliged) to find or
retrieve in the underlying Proteomics sources.
Therefore, the first initiative consists of determining
the part of the query that cannot be answered by
available proteomics sources.
4.1 Query Rewriting Formalization
We represent by the following couple
),'(' mappingsMproteomicsOOSch = , the set of semantic
knowledges about our domain of interests, which is
proteomics.
The concepts annotations, defined
in
proteomicsO' will serve to enrich Global query
before rewriting process. The semantic mappings
will show query answering capabilities of the
underlying sources.
Given a Global query
Q and the knowledges
couple
O'Sch , our rewriting approach consist to
determine two sub-queries
valide
Q and
invalide
Q .
Explicitly, we shall calculate:
invalide
Q''Q = having a size as minimal as
possible. The Size of a query is the number of
atomic goals that it contains. Sub-Query
''Q
cannot be answered by underlying sources, at
the moment of the sending of the Global
query
Q . This initial operation has the role of
cleaning up
Q
of domain concepts/properties
which are not yet available, as Web
proteomics registered resources. So, no
processing will be realized on
''Q , in the
future.
valide
Q'Q
=
is the part of Q that will be
rewrite using semantic mappings
M of
O'Sch . Sub-query 'Q can be answered by
registered sources. Our final goal is to propose
an intelligent subdivision of
valide
Q'Q = into
sub-queries
1
'Q ,
2
'Q ,…,
m
'Q with nm1 ≤≤ ,
n
is the number of sources available in the
integration while
m denotes the number
sources which are necessary to provide an
answer to the query
Q . So, we might find the
set
{
}
)m,'Q('Q
jj
=
of couples
)m,'Q(
jj
such
as
j
'Q
be an atomic subdivision of 'Q that
will be answered by mapping
j
m
.
We can easily remark that several rewritings can
be proposed, we will call them Candidate rewritings.
In fact, more than one source, and so mapping, could
provide the same resources searched.
The algorithm receives as input a global
query
Q
, a schema O'Sch and generate as output all
candidates’ rewritings
)Q(r
i
.
4.2 Hypergraph-based Algorithm
In practice, Global queries are expressed like
conjunctive queries using Trees-Logics. So, a
rewriting
'Q
is a suitable conjunction of constraints.
These constraints might be checked by all final
answers of the global query
Q , because they
constitute an indication of resources that may be
retrieved from adequate sources.
In order to provide a characterization of our
query rewriting problem, we give an alternative
formulation of the rewriting formalization.
Given a Global Query
Q and the semantic
knowledges couple
O'Sch , query rewriting consists
to compute two sub queries
valide
Q and
invalide
Q on
the basis of mappings set
M
, such as:
invalidevalide
QQQ
∧
=
(1)
We are searching for all candidates rewritings,
formulated as the conjunction of constraints:
i
m
1ivalide
C'QQ
=
==
(2)
All constraints
i
C are logical representation of the
user specific needs. Finally, our motivation is to
answer the fundamental question which is to find,
given
Q a new query called rewriting expressed by
ICEIS 2009 - International Conference on Enterprise Information Systems
126

i
m
1ivalide
C'QQ
=
==
such as
'Q
denotes as much as
possible the resources expected by query
Q ?
We have said that several and alternatives
rewritings are possible, due to the fact that more
than one mapping could be used to reformulate an
atomic goal of the Global query. From this point of
view, rewriting problem which requires generating
all candidates’ rewritings can be characterized as a
current Hypergraph Theory problem of computing
all minimals Transversals of an Hypergraph.
Generate a Transversal Hypergraph consists of
generate all minimals Transversals.
4.2.1 Definition of Hypergraph (Kavvadias,
2005)
An Hypergraph
H
is an ordered pair ),( EVH
=
where
{}
n21
v,...,v,vV = is a finite set of elements
and
{}
m21
E,...,E,EE = is a family of subsets of
V
such that
-
i
E )m,...,1i(, =
-
VE
m
1i
i
=
=
∪
The elements of
V are called nodes while the
elements of
E are called hyperedges of the
hypergraph
H
. A hypergraph can be seen as a
generalization of a graph where the restriction of an
edge having only two nodes does not hold.
4.2.2 Definition of Transversals (Kavvadias,
2005)
Let )E,V(H = be an hypergraph. A set VT ⊆ is
called a Transversal of
H
if it intersects all its
hyperedges, i.e,
i
ET EE,
i
. A transversal
T
is called minimal if no proper subset '
T
of T is a
transversal of
H
.
The Transversal Hypergraph
)H(Tr of an
hypergraph
H
is the family of all minimal
transversals of
H
.
From a rewriting query problem, we need to give
a mathematical characterization, by defining an
associated Hypergraph
)E,V(H
M,Q
, built as
follows:
For every mapping
i
m , describing a local
concept from
M
, as a logical function of
O’proteomics global concepts, we associate a
vertice
i
m
V in the hypergraph
)E,V(H
M,Q
and
[]
{
}
n,1i,VV
i
m
=
.
For every constraint
i
C of the Global query Q ,
we associate an hyperedge
i
C
E in the
hypergraph
),(
,
EVH
MQ
. To simplify, we
suppose that all these constraints are
describing atomics goals. So, each
hyperedge
i
C
E is a set of mappings, calculated
by considering those mappings which are
relevant to answer these goals.
A classical algorithm to compute minimal
Tranversals of an hypergraph is proposed and
available in (Mannila, 1994). Many papers (Eiter,
1995) have discussed about algorithm of generation
of Hypergraph Transversal, which is a set of
minimal Transversals. One of the first results
remains Berge’s Algorithm (Berge, 1989), but
several variants have been proposed in order to deal
with the algorithm complexity (Rey, 2003).
Now, we present our Query rewriting algorithm
called Q-Candidates’Finder, which integrates the
better and efficient complexity of the classical
Algorithm:
Q-Candidates’Finder Algorithm.
Input: A Query Q and
mappings)M,proteomics(O'OSch' =
Output:The set
o
f candidates rewriting such
as
{
}
)
invalide
Q,
valide
Q(
candidates
Q =
1: Build the associated Hypergraph
),(
,
EVH
MQ
2: Compute
i
k
1iinvalide
CQ
=
= such as
i
C
is not
provided by any mappings in
M .
3: Build the associated Hypergraph
)E,V(H
*
M,Q
*
4:
=
candidates
Q
5: Generate the Hypergraph Transversal
of
)E,V(H
*
M,Q
*
- Let be HypTransv - Using the Classical
Algorithm [Mannila, 1994]
6: For all edge
{
}
HypTransvV,...,V,VX
p21
mmm
=
do
7:
[]
{
}
pjmQQQrQ
jjvalide
,1),,'(')( ∈===
where
j
'Q is a subdivision of
valide
Q
that will be answered by the mapping
j
m
.
8:
validecandidatescandidates
QQQ ∪
=
9: End For
10 : Return
candidates
Q
AN USER-CENTRIC AND SEMANTIC-DRIVEN QUERY REWRITING OVER PROTEOMICS XML SOURCES
127
4.3 Q-Candidates’ Finder Illustration
To illustrate the proposed rewriting approach, let us
consider the following mappings (L.A.V approach).
As the domain Ontology is characterized as a Tree,
semantic mappings might express subsomption
(Sub) and equivalence (Eq) relations that exist
between Local XML Schemas, also abstracted as
Trees, and the Ontology’s Tree model. We suppose
in order to simplify this illustration that we have
only simple paths mappings (and so, no sub-Trees)
between Concepts, according to 1:1 cardinality. For
every registerd proteomics’source, we provide the
LHS (Left Hand Side expressing Local
Concepts/Properties), the TYPE (the type of
mappings), and the RHS (Right Hand Side,
expressing Ontology Concepts/Properties) of the
current mapping.
Table 1: Mapping Table.
LHS TYPE RHS
O’proteomics
Ontology
Gene (Genes, Proteins, Species, Organisms)
Proteine (IdProteins, Peptides, DevStadium)
…
Mapping m1:
Description Of
Source 1
S1_Gene (
S1_GeneName,
S1_ProteinName;
S1_species,
S1_organisms)
Eq
…
Gene (
Genes,
Proteins,
Species,
Organism)
Mapping m2:
Description Of
Source 2
S2_Gene (
S2_NomGenes,
S2_NomProteine,
S2_Especes,
S2_Organismes)
Eq
….
Gene (
Genes
Proteins
Species
Organisms)
Mapping m3:
Description Of
Source 3
S3_TreeLife (
S3_Species,
S3_Genus…)
Sub
…
Gene (
Species,
Organisms)
We have just shown mappings which are
relevant for the Query we shall process. Note once
again, that we are considering corresponding paths
of these Concepts/Properties in their abstract Trees.
According to this mapping table, we can say that
the ontology includes Concept Gene and Proteine
with their relative Properties such as Genes,
Proteins, and Species … Mappings m1 and m2 show
that Source I and Source II provide the properties
such as Gene, Proteins, Species, and Organisms,
while the mapping m3 illustrate that Source III only
provides properties such as Species and Organisms.
Let us consider now the following query which is
expressed over the domain ontology, O’proteomics :
What are the genes which proteins could have a
peptide Signal and for which, it is assumed that they
are expressed at Tardive Shizont stadium for the
Plasmodium falciparum?
4.3.1 Hypergraph Construction
Intuitively,
Q
can be expressed like a conjunction of
the following constraints:
SpeciesOrganisms
sDevStadiumPeptidesGenes
CC
CCCQ
C
Proteins
∧
=
(3)
In practice, the user will formulate this request
by using an user-friendly graphic interface, and the
generation of its Trees-Logics version is done
automatically. He will choose the Concept Genes
and indicate for each property Gene, Proteins,
Species, Organisms, Peptides, DevStadiums the
expected values.
The associated hypergraph
),(
,
EVH
MQ
consists
of the following sets of vertices and edges:
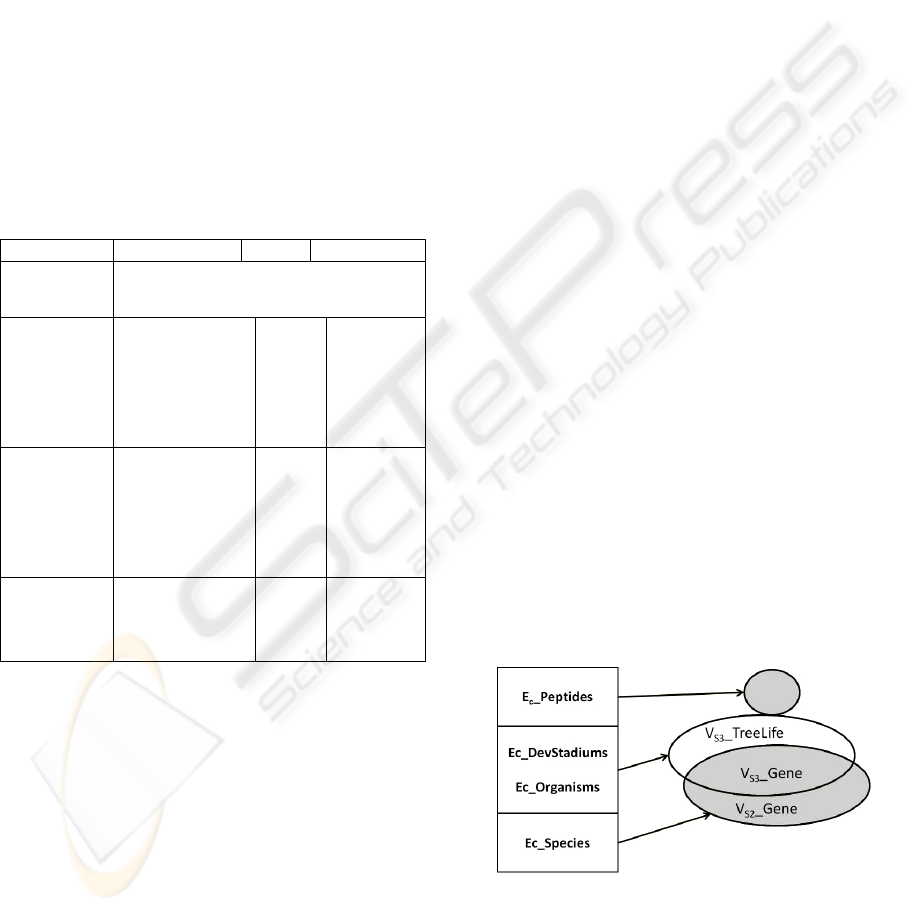
{
}
,,
3_1_2 TreeLifeSGeneSGeneS
VVVV
−
=
(4)
and
,,,
,
__Pr_
__,_
EE E
E
EEE
C Genes C oteins C Peptides
C DevStadiums C organisms C Species
⎧
⎫
⎪
⎪
=
⎨
⎬
⎪
⎪
⎩⎭
(5)
We could see this illustration using a
Sets’Theory point of view. We materialize all query
constraints as Sets that contain the providers’
mappings.
We show graphically these sets of mappings:
Figure 1: Associated Hypergraph of the illustration.
4.3.2 Determination of
invalide
Q
invalide
Q is the part of Q that cannot be answered by
available sources. That means, we might constitute
ICEIS 2009 - International Conference on Enterprise Information Systems
128

invalide
Q with all Q ’s constraints, which are
characterized by empty hyperedges of
hypergraph
),(
,
EVH
MQ
.
We can easily see that no mapping provides the
hyperedges DevStadiums and Peptides. These
hyperedges are empty of associated mappings (Q-
Candidates’Finder: Line 2).
Hence:
sDevStadiumPeptidesinvalide
CCQ =
(6)
4.3.3 Determination of
valide
Q
valide
Q is the part of Q that can be answered by
available sources. That means, we might constitute
valide
Q with all
Q
’s constraints, which are
characterized by non-empty hyperedges of
hypergraph
),(
,
EVH
MQ
.
So, we have:
SpeciesOrganismsGenesvalide
CCCQ C
Proteins
∧=
(7)
In fact,
valide
Q
means that we could only answer
the following request, given semantic mappings:
Give the genes which code all proteins, for the
Plasmodium falciparum?
4.3.4 Determination of
candidates
Q
From calculated above, we generate associated
hypergraph Hypergraph
),(
*
,
*
EVH
MQ
, (see Line 3).
Intuitively, according to Sets’Theory vision,
finding all candidates rewriting suppose firstly to
construct the Cartesian product of all sets of
mappings. It is obvious that Elements of the
Cartesian product are 4-uplets in our illustrative
example. So, we must generate for each 4-uplet, an
associated Set (These sets correspond to
Transversals). In fact, it will be useful to use a
minimal number of Sources that would be requested.
This condition is guaranteed if we consider only
associated sets that not contain another associated
Set (These sets are minimals Transversals). That is
why we call the Classical Algorithm (see Line 5).
The maximal cardinality of our example
Transversals is 4, it is the size of
valide
Q .
From Sets’Theory point of view we can say that
any set that contain
GeneS
V
_1
and
GeneS
V
_2
is a
Transversal and constitutes possible rewritings of
Q .
The minimal Transversal are
{}
GeneS
V
_1
and
{
}
GeneS
V
_1
constitute two candidates rewritings.
In our case, we could find 36 4-uplets, associated
with 6 Transversals but only 2 are minimal
Transversals.
5 CONCLUSIONS
This paper deals with Global query rewriting, which
consists in data integration context, to rewrite the
global query expressed in terms of concepts and
their properties defined in global schema domain
ontology) into suitable terms of local data sources.
The Query rewriting process is based on semantic
mappings. Our knowledge domain concern
Proteomics research, and so we have proposed
ontology according to interviews and talks with
biologists and bio-informaticians, called
O’proteomics. We provide a characterization of the
Query rewriting problem based on Hypergraph
Theory. We have presented the classiscal algorithm
that computes all minimals Tranvsersals, given an
Hypergraph. We observe that those minimals
Transversals correspond to Candidates rewritings of
the Global Query.
The formalization and the specification of the
proteomics semantic knowledges and some efforts to
find automatically mappings between the ontology
and the different local Schemas. Therefore, we need
to better defined a logical formalisme or language to
specify synatx and semantics data Trees. It could be
seen as a subset of Description Logics or based on
Psi-terms formalism. After this essential choice, we
will try to provide a prototype.
This paper shows briefly our current research
that aims to provide a semantic framework to realize
a Data Integration over XML bio-Sources on the
Web. We will define some relevant criteria to rank
candidates rewritings, necessary to select an optimal
and qualitative rewriting
optimal
Q . An efficient way
for selecting best rewritings, iteration by iteration,
will permit us to investigate the properties and the
optimiwation of our algorithm. These relevant
criteria could concern user preferences, quality of
underlying sources, etc…
REFERENCES
Amann, B., Beeri C., Fundulaki I., Scholl, M., 2002.
Ontology-based integration of XML web resources. In
Proceedings of International Semantic Web
Conference '02
, Pages: 117-131.
AN USER-CENTRIC AND SEMANTIC-DRIVEN QUERY REWRITING OVER PROTEOMICS XML SOURCES
129

Baader, F., Calvanese, D., McGuinness, D., Nardi, E.D.,
Patel-Schneider, P., 2003.
The Description Logic
Handbook, Theory, Implementation and Applications
,
Cambridge University Press, Cambridge.
Benatallah, B., Hacid M-S., Paik H-y., Rey C., Toumani
F., 2006. Towards semantic-driven, flexible and
scalable framework for peering and querying e-catalog
communities, In
Elsevier’s Journal of Information
Systems
, Pages: 266-294.
Berge, C., 1989.
Hypergraphs. North Holland,
Amsterdam, ISBN 0 444 874895; QA166.23.B4813.
Bishop, M., 1999. Genetics Databases, Academic Press.
Bray, T., Paoli, J., Sperberg-McQueen, 1998. “
Extensible
Markup Language (XML) 1.0
,” W3C February
Recommendation, available online at
http://www.w3.org/TR/REC-xml.
Davidson, S.B., Overton, C., Buneman, P., 1995.
Challenges in integrating biological data sources.
Journal of Computational Biology, 2(4):557–572.
Delobel, C., Reynaud, C., Rousset, M-C., Sirot, J-P.,
Vodislav, D., 2003. Semantic integration in Xyleme: a
uniform tree-based approach, In
Elsevier’s Journal of
Data & Knowledge Engineering
44, Pages: 267–298.
Deutsch, A., Tannen, V., 2003. Reformulation of XML
Queries and Constraints, In Proceedings of the 9th
International Conference on Database Theory (ICDT)
,
Pages 225-241.
Deutsch, A., Tannen, V., 2005. XML queries and
constraints, containment and reformulation,
Elsevier’s
Journal of Theoretical Computer Science
, 336 Pages:
57-87
Eiter, T., Gottlob, G., 1995. Identifying the minimal
transversals of a hypergraph and related problems.
SIAM Journal on Computing, 24(6), Pages: 1278-
1304.
Fundulaki, I., Amann, B., Beeri, C., Scholl, M., 2002.
STYX: Connecting the XML World to the World of
Semantics Web resources. In Proceedings of EDBT'
2002, Prague, Czech Republic
.
Gottlob, G., Koch, C., Schulz, K.U, 2004. Conjunctive
Queries over Trees, In Proceedings 23rd ACM
SIGMOD-SIGART Symposium on Principles of
Database Systems (PODS 2004)
, Paris, France. ACM
Press, New York, USA, Pages: 189-200.
Halevy, A., 2001. Answering queries using views: a
survey, In
Proceedings of Very Large Data Bases. 10
(4), Pages: 270–294.
Halevy, A., Ives Z., Tatarinov I., Mork P., 2003. Piazza:
Data management infrastructure for semantic web
applications
. In Proceedings of the International
World Wide Web Conference
.
Kavvadias, D., Stavropoulos, E., 2005. An Efficient
Algorithm for The Transversal Hypergraph
Generation, In
Journal of Graph Algorithms and
Applications
, Vol.9, No.2, Pages: 239-264.
Kohler, J., 2004. Integration of Life Science databases, In
Elsevier's Drug Discovery Today Journal,
BIOSILICO Vol.2, No.2.
Lehti, P., Fankhauser, P., 2004. XML Data Integration
with OWL: Experiences and Challenges, In
Proceedings of Symposium on Applications and the
Internet (SAINT'04), Pages: 160 -170.
Levy A., Rajaraman A., Ordille J., 1996. Querying
Heterogeneous Information Sources Using Source
Description
s, In Proceedings of Very Large Data
Bases Conference
, pages 251-262, Mumbai, India.
Mannila, H., Raiha, K-J, 1994.
The Design of Relational
Databases
. Addison -Wesley, Wokingham, England.
Manolescu, I., Florescu, D., Kossmann, D.K, 2001.
Answering XML queries over heterogeneous data
sources. In
Proceedings of the 27th International
Conference on Very Large Data Bases (VLDB ’01)
,
Orlando, Pages: 241–250.
Rey, C., Toumani, F., Hacid, M.-S., Leger, A., 2003,
An
algorithm and a prototype for the dynamic discovery
of e-services, Technical Report, LIMOS, Clemont-
Ferrand, France.
Schlieder, T., 2001. ApproXQL: Design and
Implementation of an Approximate Pattern Matching
Language for XML, Technical Report, Freie
Universitat Berlin.
Thompson, H.S., 2000. “XML Schema Part 1: Structures,”
W3C, work-in-progress, current as of Apr. 2000.
ICEIS 2009 - International Conference on Enterprise Information Systems
130
