
Stability of Ensemble Feature Selection on High-Dimension and
Low-Sample Size Data
Influence of the Aggregation Method
David Dernoncourt
1,2,3
, Blaise Hanczar
4
and Jean-Daniel Zucker
1,2,3,5
1
Institut National de la Sant´e et de la Recherche M´edicale, U872, Nutriomique,
´
Equipe 7,
Centre de Recherches des Cordeliers, Paris 75006, France
2
Universit´e Pierre et Marie-Curie - Paris 6, Nutriomique, 15 rue de l’
´
Ecole de M´edecine, Paris 75006, France
3
Institute of Cardiometabolism and Nutrition, Assistance Publique-Hˆopitaux de Paris, CRNH-
ˆ
Ile de France,
Piti´e-Salpˆetri`ere, Boulevard de l’Hˆopital, Paris 75013, France
4
LIPADE, Universit´e Paris Descartes, 45 rue des Saint-P`eres, Paris, F-75006, France
5
Institut de Recherche pour le D´eveloppement, IRD, UMI 209, UMMISCO, France Nord, F-93143, Bondy, France
Keywords:
Feature Selection, Stability, Ensemble, Small Sample.
Abstract:
Feature selection is an important step when building a classifier. However, the feature selection tends to be
unstable on high-dimension and small-sample size data. This instability reduces the usefulness of selected
features for knowledge discovery: if the selected feature subset is not robust, domain experts can have little
trust that they are relevant. A growing number of studies deal with feature selection stability. Based on the
idea that ensemble methods are commonly used to improve classifiers accuracy and stability, some works
focused on the stability of ensemble feature selection methods. So far, they obtained mixed results, and as far
as we know no study extensively studied how the choice of the aggregation method influences the stability of
ensemble feature selection. This is what we study in this preliminary work. We first present some aggregation
methods, then we study the stability of ensemble feature selection based on them, on both artificial and real
data, as well as the resulting classification performance.
1 INTRODUCTION
Feature selection is a critical step of the supervised
classification procedure and specially in the small-
sample high dimension settings. Small-sample high
dimension settings refers to problems where the num-
ber of features is higher than the number of examples.
This kind of problem is increasingly frequent, espe-
cially in bioinformatics with the massive production
of ”omics” data. In this context the learning algo-
rithms met several problems called the curse of di-
mensionality (Simon, 2003). In high dimension, find-
ing the actual informative features becomes more dif-
ficult and the risk of overfiting strongly increases. The
consequence is the worse generalization performance
of the classifiers (Jain and Chandrasekaran, 1982).
Secondly, very high dimension alone is an issue it-
self, as classifiers frequently do not scale well to huge
numbers of features, leading to increased computa-
tion times. Thirdly, a classifier based on a small sub-
set of genes will be easier and less expensive to use in
practice. Moreover, a classifier based on a high num-
ber of features will not be easily interpretable. The
point problem is about the robustness of the selected
features. To obtain a confident classifier, the selected
subset has to be stable. To deal with all of these prob-
lems a feature selection is necessary in order to reduce
dimensionality of the data.
Feature selection refers to the process of remov-
ing irrelevant or redundant features (in our con-
text, genes) from the original set of features F =
{ f
1
, f
2
,..., f
|F |=D
}, so as to retain a subset S ⊂ F con-
taining only informative features useful for classifi-
cation (Liu et al., 2010). Beyond classification per-
formance, the other main objective of the gene selec-
tion is to obtain a reliable and robust list of predictive
genes: a gene which is regularly selected over several
datasets dealing with the same problem – or at least
over various random subsamples of the same dataset
– is more likely to be really relevant, and is of greater
interest to domain experts willing to use the classi-
fication results for knowledge discovery purposes. In
325
Dernoncourt D., Hanczar B. and Zucker J..
Stability of Ensemble Feature Selection on High-Dimension and Low-Sample Size Data - Influence of the Aggregation Method.
DOI: 10.5220/0004922203250330
In Proceedings of the 3rd International Conference on Pattern Recognition Applications and Methods (ICPRAM-2014), pages 325-330
ISBN: 978-989-758-018-5
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

practice, this is generally not the case, and many stud-
ies on feature selection stability emphasized the diffi-
culty to obtain a reproducible gene signature on high-
dimension small-sample data (Ein-Dor et al., 2006;
Haury et al., 2011). Stability of feature selection
strongly depends on the N/D ratio and problem dif-
ficulty. For example in one previous work we have
shown that on a simple two Gaussian classes problem
with 1000 features, a t-test based selection has a prob-
ability of more than 0.95 to select the very informative
features when N = 1000, but this probability falls to
less than 0.2 when N = 50 (Dernoncourt et al., 2014).
A growing number of studies have been dealing
with feature selection stability, be it either to intro-
duce stability measures (Kuncheva, 2007; Somol and
Novoviˇcov´a, 2010), to compare the stability of exist-
ing methods (Haury et al., 2011), and/or to propose
innovative, stability-focused feature selection meth-
ods (Saeys et al., 2008; Han and Yu, 2012). Some
works, such as (Somol et al., 2009; Abeel et al., 2010;
Haury et al., 2011), also investigated the stability of
ensemble feature selection methods. The main idea
behind ensemble methods is to produce several indi-
vidual feature selections and combine them in order
to obtain a selection that outperforms every one of
them. This idea is based on the concept of ”wisdom of
the crowd”, which states that under certain controlled
conditions, the aggregation of information from sev-
eral sources, results in decisions that are often su-
perior to those that could have been made by any
single individual - even experts (Surowiecki, 2004;
Rokach, 2010). In practice, ensemble methods have
been widely applied to classifiers and since they can
improve classifiers accuracy and stability, we can sup-
pose that they should provide similar benefits to fea-
ture selection techniques (Yang et al., 2010). So far,
works on ensemble feature selection have mainly fo-
cused on classification accuracy, showing accuracy
gains (or losses) to be problem-dependent (Han et al.,
2013) and filter-dependent (Wald et al., 2013). Works
which also studied stability have obtained mixed re-
sults too, leaving the general impression that both sta-
bility and accuracy gains (or losses) from ensemble
methods are problem-dependent (Saeys et al., 2008).
However, those works have often measured stabil-
ity over overlapping resamplings, which strongly in-
creases the measured stability (Haury et al., 2011),
and might also impact stability variations, and as far
as we know no study extensively studied how the
choice of the aggregation method influences the sta-
bility of ensemble feature selection methods.
In this preliminary work, we start investigating
how ensemble methods improve feature selection
(FS) stability, with a focus on the impact of the aggre-
gation method. We first briefly present the stability
measures and the three ensemble aggregation meth-
ods we used. Then we perform an empirical analysis
of feature selection stability on both artificial and real
microarray datasets.
2 ENSEMBLE FEATURE
SELECTION
Creating an ensemble feature selection can be divided
into two steps. The first step is to create a set of di-
verse feature selectors. The second step is to aggre-
gate them.
2.1 Diversity Generation
The diversity of feature selectors is a crucial condi-
tion for obtaining a ”wise crowd” (Surowiecki, 2004),
necessary for an efficient ensemble. It can be obtained
via different methods such as:
• manipulating the training sample: typically, re-
sampling the training set so as to perform each FS
of the ensemble on a different training set,
• manipulating the FS method: for instance, use dif-
ferent parameters for each FS, if the FS method
has parameters,
• partitioning the search space: each FS is per-
formed on a different search space, for instance,
random forest learns each tree on a random, dif-
ferent, small subset of features,
• hybridization: use several FS methods in the en-
semble,
or a combination of those (Rokach, 2010). In this pa-
per, we focused on manipulating the training sample,
which is the most commonly used method, and ob-
tained diversity by bootstrapping the training samples
B = 40 times, based on previous works that showed
that ensemble FS doesn’t improve much when in-
creasing the amount of bootstrap samples further than
40 (Abeel et al., 2010) or even 20 (Saeys et al., 2008).
2.2 Aggregation
We tested the following methods of aggregation:
• Average score: on each bootstrap sample, the
FS method outputs a score s
f
i
, j
for each gene f
i
.
We simply average the score of a gene over the
bootstrap samples in order to obtain the ensemble
score W
f
i
:
W
f
i
=
∑
B
j=1
s
f
i
, j
B
(1)
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
326

• Average rank (Abeel et al., 2010): on each boot-
strap sample, the score of each gene s
f
i
, j
is con-
verted into a rank r
f
i
, j
. Then we average the rank
of a gene over the bootstrap samples in order to
obtain the ensemble score:
W
f
i
=
∑
B
j=1
r
f
i
, j
B
(2)
• Stability selection (Meinshausen and Bhlmann,
2010): the ensemble score W
f
i
of each gene is ob-
tained by measuring how often the gene ranks in
the top d of each bootstrap sample:
W
f
i
=
∑
B
j=1
I( f
i
, j)
B
(3)
where I( f
i
, j) = 1 if gene f
i
is ranked in the top d
on FS performed on the j
th
bootstrap sample, and
I( f
i
, j) = 0 otherwise.
Finally, for each aggregation strategy, the d genes
with the largest score W
f
i
are retained as the final en-
semble selection.
3 EXPERIMENTAL DESIGN
We performed experiments on both artificial and real
data in order to assess the impact of ensemble meth-
ods on the stability of feature selection. We used four
base FS methods: t-score, random forest, recursive
feature elimination using support vector machines
(SVM-RFE), and mutual information. For each of
those methods, feature selection was performed with
and without the ensemble aggregation methods de-
scribed in the previous section. We then measured
the stability of the feature selection on strictly non-
overlapping sets (different generated sets on artificial
data, resamplings on real data), and the average classi-
fication error rate obtained with a linear discriminant
analysis (LDA) classifier on the test sets.
3.1 Stability Measure
Many different measures have been described to mea-
sure stability. As our main stability measure, we
chose to use the relative weighted consistency (Somol
and Novoviˇcov´a, 2010), CW
rel
, which evaluates how
frequently each feature is selected, among the features
which have been selected at least once. We chose it
because it ignores the stability of non-selected fea-
tures (which would artificially increase the stability
measure on large datasets where many features are
irrelevant and easy to exclude), and because it’s ad-
justed to take into account the proportion of overlap-
ping features due to chance.
Let S = {S
1
,S
2
,...,S
ω
} be a system of ω gene sub-
sets obtained from ω runs of the feature selection rou-
tine on different samplings, Ω =
∑
ω
i=1
|S
i
| be the total
number of occurrences of any gene in S and F
f
be the
number of occurrences of gene f ∈ F in system S.
The weighted consistency CW was defined as follow:
CW(S) =
∑
f∈X
F
f
Ω
·
F
f
− 1
ω− 1
(4)
and CW
rel
is obtained by adjusting CW on its minimal
and maximal possible values CW
min
and CW
max
:
CW
rel
(S, F ) =
CW(S) −CW
min
(Ω,ω,F )
CW
max
(Ω,ω) −CW
min
(Ω,ω,F )
(5)
3.2 Artificial Data
We used three different artificial data structures, all
based on a two-classes Gaussian model with D =
1000 genes. Each of the two classes follows a nor-
mal distribution defined respectively by N (µ,I) and
N (−µ,Σ), where µ is a vector of means such that
|µ| = D and Σ is the covariance matrix.
In the first data structure, NC100, all genes are in-
dependent (Σ is the identity matrix) and the elements
µ
i
of µ consisted of d = 100 elements µ
i
= 1 (genes
useful for classification) and D − d = 900 elements
µ
i
= 0 (noise genes). Then µ was scaled down so that
F would yield a specified Bayes error (ε
Bayes
= 0.10).
In the second data structure, NC, all genes are in-
dependent and the elements µ
i
of µ were drawn from
a triangular distribution with a lower limit and mode
equal to 0 (probability density function: f(x) = 2−2x
for x ∈ [0;1]). In order to obtain a more realistic prob-
ability density, we then raised µ to a power of γ = 2,
similarly to the method used in (Dernoncourt et al.,
2014). Again, µ was scaled down so that F would
yield ε
Bayes
= 0.10.
In the third data structure, CB, we added correla-
tions within blocks of ten genes, by using the covari-
ance matrix
Σ =
Σ
1
0 ··· 0
0 Σ
2
··· 0
.
.
.
.
.
.
.
.
.
.
.
.
0 0 ··· Σ
100
, (6)
where Σ is a block diagonal matrix and Σ
i
is a 10× 10
square matrix with elements 1 along its diagonal and
0.5 off its diagonal, similarly to the method used in
(Han and Yu, 2012). We used the same µ as in the NC
dataset.
From these models, 50 training sets were gener-
ated, on which the FS methods were performed and an
LDA classifier was trained. Each classifier was then
applied to a test set consisting of 10000 instances to
estimate its error rate.
StabilityofEnsembleFeatureSelectiononHigh-DimensionandLow-SampleSizeData-InfluenceoftheAggregation
Method
327

Table 1: Microarray datasets.
Name N D Source
Colon cancer 62 2000 Alon et al., 1999
Leukemia 72 7129 Golub et al., 1999
Breast cancer Pawitan 159 8112 Pawitan et al., 2005
Lung cancer 203 2000 Bhattacharjee et al., 2001
Breast cancer Vijver 294 2000 van de Vijver et al., 2002
3.3 Real Data
We experimented with five publicly available mi-
croarray datasets, listed in Table 1. For each dataset,
50 training sets were generated by randomly drawing
half instances from the dataset (without replacement).
For each of them, feature selection was performed and
a classifier was trained (using the same methods as
with the artificial data). Each classifier was then ap-
plied on a test set consisting of the samples not in-
cluded in the corresponding training set. Stability of
the feature selection was measured within each pair
of training and test sets, so as to have no overlap. The
final measure of stability corresponds to the average
of those 50 measures.
4 RESULTS
4.1 Artificial Data
Table 2 presents the stability of feature selections and
the error rate of resulting classifiers on the artificial
datasets. In general, we observed that ensemble fea-
ture selection providessimilarly or more stable results
than non-ensemble (single) feature selection, and a
similar or lower error rate.
T-score obtained the highest stability and lowest
classification error rate overall. It further benefited
from its ensemble version, but only with the average
score aggregation: the average rank and stability se-
lection aggregation slightly degraded its stability and
did not improve the classification error rate.
Compared to t-score, SVM-RFE had a slightly
lower stability on non-correlated data and half the sta-
bility on correlated data, but with a similar error rate.
It did not benefit from the ensemble version on non-
correlated data, but stability was improved on corre-
lated data, similarly with all aggregation types.
Random forest was the worst performer, with half
the stability of t-score on all datasets, and a higher er-
ror rate on the uncorrelated datasets. It was however
much more stable in its ensemble version, reaching
similar stability levels and error rate as single SVM-
RFE on non-correlated data, and a higher stability
(and somewhat lower error rate) than ensemble SVM-
RFE on correlated data. As with SVM-RFE, the ag-
gregation method did not matter here.
4.2 Real Data
Table 3 presents the stability of feature selections and
the error rate of resulting classifiers on the real mi-
croarray datasets. In general, similarly to the artifi-
cial data, we observed that ensemble feature selection
provides similarly or more stable results than single
feature selection, with the exception of t-score on the
leukemia dataset. Unlike what we observed on arti-
ficial data though, the error rate was in some cases
increased by the ensemble selection.
T-score obtained the highest stability on 2 out of
5 datasets. In 4 datasets, its ensemble version had
an improved stability, but only with the average score
aggregation, and in half cases (colon cancer and lung
cancer datasets) at the cost of a largely increased er-
ror rate. On the leukemia dataset, ensemble methods
reduced stability and increased error rate.
SVM-RFE obtained the highest stability on 3 out
of 5 datasets. In all datasets, its ensemble version had
an improved stability, the best improvement occurring
with the average score aggregation, closely followed
by stability selection. Aggregation by average rank
did not perform as well: it providedthe worse stability
increase on the colon cancer and lung cancer dataset,
and no stability increase or even a stability degrada-
tion on the other datasets. The error rate was generally
unchanged by ensemble methods, except for a 10%
increase in the colon cancer dataset (with any aggre-
gation method) and a 20% decrease in the lung cancer
dataset (aggregation by average rank only).
Random forest had the lowest stability on 3 out of
5 datasets, and tied with mutual information on the
Vijver dataset. However it had a competitive error
rate (best without ensemble on Vijver and leukemia
datasets, best with ensemble on the colon cancer
dataset), even though the differences in error rates
between the different feature selection methods were
generally small, except on the Pawitan dataset. Sim-
ilarly to SVM-RFE, its stability was generally in-
creased by ensemble methods. Aggregation by av-
erage score and stability selection improved stability
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
328
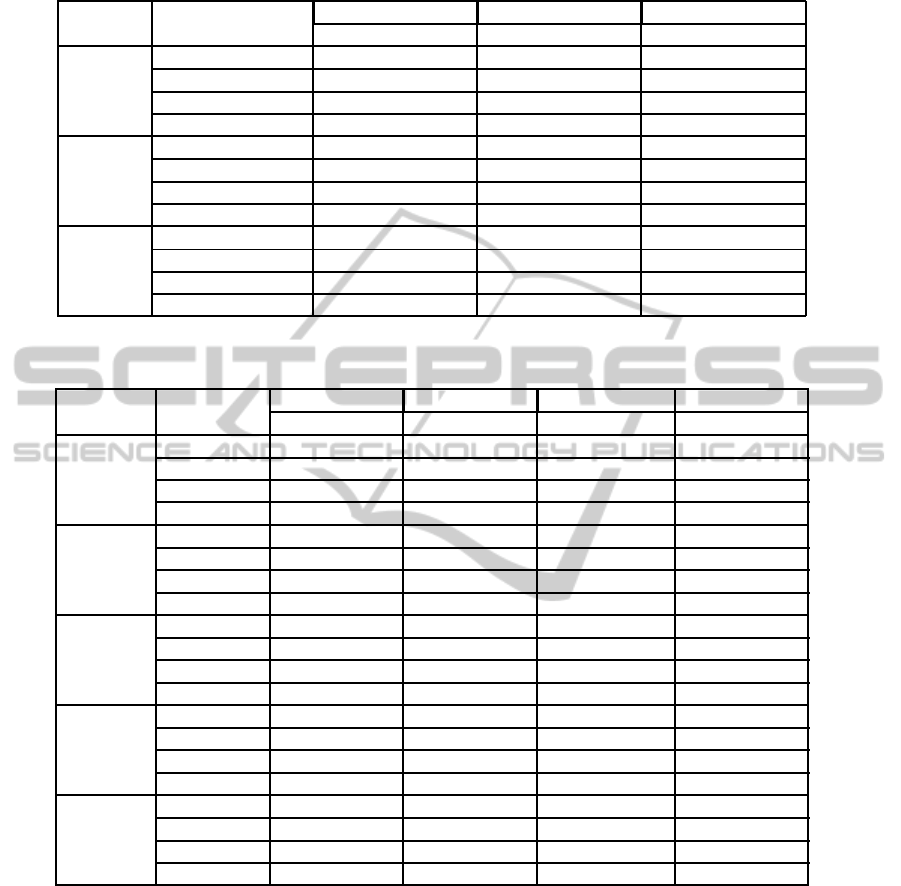
Table 2: Classification error rate and selection stability on the artificial datasets, with feature selection without ensemble and
with three ensemble aggregation methods.
Artificial Ensemble t-score SVM-RFE Random forest
data type aggregation Error rate CW
rel
Error rate CW
rel
Error rate CW
rel
NC100 Single 0.338 0.075 0.345 0.065 0.383 0.031
Average score 0.290 0.135 0.339 0.071 0.347 0.059
Average rank 0.340 0.072 0.340 0.072 0.347 0.061
Stability selection 0.339 0.071 0.344 0.068 0.345 0.062
NC Single 0.360 0.050 0.360 0.049 0.391 0.027
Average score 0.320 0.071 0.364 0.045 0.365 0.042
Average rank 0.361 0.045 0.364 0.044 0.363 0.045
Stability selection 0.366 0.043 0.363 0.045 0.368 0.040
CB Single 0.239 0.235 0.242 0.113 0.244 0.099
Average score 0.223 0.265 0.238 0.152 0.232 0.193
Average rank 0.235 0.226 0.238 0.154 0.230 0.195
Stability selection 0.237 0.217 0.239 0.150 0.233 0.190
Table 3: Classification error rate and selection stability on the microarray datasets, with feature selection without ensemble
and with three ensemble aggregation methods.
Data Ensemble t-score SVM-RFE Random forest Mutual inf
aggregation Error CW
rel
Error CW
rel
Error CW
rel
Error CW
rel
Colon Single 0.188 0.310 0.182 0.448 0.196 0.163 0.181 0.140
Score 0.305 0.327 0.203 0.588 0.179 0.206 0.217 0.149
Rank 0.215 0.277 0.203 0.494 0.199 0.163 0.209 0.149
Stability 0.210 0.262 0.206 0.568 0.177 0.210 0.213 0.145
Leukemia Single 0.049 0.322 0.044 0.525 0.042 0.220 0.050 0.263
Score 0.117 0.315 0.051 0.581 0.043 0.265 0.052 0.300
Rank 0.054 0.269 0.047 0.517 0.067 0.094 0.053 0.297
Stability 0.058 0.246 0.047 0.565 0.046 0.269 0.049 0.294
Pawitan Single 0.342 0.071 0.283 0.129 0.309 0.011 0.313 0.023
Score 0.328 0.085 0.283 0.180 0.320 0.012 0.343 0.024
Rank 0.344 0.065 0.298 0.095 0.324 0.015 0.335 0.024
Stability 0.317 0.047 0.289 0.180 0.314 0.011 0.329 0.022
Lung Single 0.054 0.515 0.084 0.377 0.060 0.342 0.061 0.372
Score 0.076 0.536 0.083 0.498 0.064 0.398 0.063 0.379
Rank 0.058 0.417 0.067 0.444 0.063 0.389 0.064 0.377
Stability 0.058 0.445 0.082 0.487 0.063 0.394 0.064 0.376
Vijver Single 0.382 0.254 0.377 0.159 0.359 0.078 0.360 0.077
Score 0.364 0.345 0.373 0.221 0.368 0.107 0.357 0.091
Rank 0.371 0.237 0.368 0.158 0.371 0.105 0.360 0.092
Stability 0.374 0.237 0.376 0.215 0.371 0.106 0.359 0.088
equally and on all datasets, except for the Pawitan
dataset. Aggregation by average rank was more in-
consistent: similar improvements as the other aggre-
gation methods on Vijver and lung cancer datasets,
a better improvement on the Pawitan dataset, where
random forest had a very low stability compared to
the other methods, no improvement compared to sin-
gle random forest on the colon cancer dataset, and an
important degradation on the leukemia dataset.
Mutual information had the lowest stability on
the colon cancer dataset, yet competitive error rates.
The ensemble versions had a similar (colon cancer,
Pawitan, and lung cancer datasets) or moderately im-
proved (leukemia and Vijver datasets) stability, with
no marked difference in favor of a specific aggrega-
tion method. Ensemble increased the error rate in
the colon cancer and Pawitan dataset, again with no
marked difference between aggregation methods.
5 CONCLUSIONS AND FUTURE
WORK
In this work, we studied how ensemble feature selec-
tion methods influence the stability of the gene selec-
StabilityofEnsembleFeatureSelectiononHigh-DimensionandLow-SampleSizeData-InfluenceoftheAggregation
Method
329

tion and, to a lesser extent, the error rate of the re-
sulting classifier on microarray data and artificial data
of similar dimension. We focused on the aggrega-
tion method used in the ensemble procedure, because
that is an important aspect of the ensemble construc-
tion procedure which, to our knowledge, had scarcely
been investigated in such a setting before. Similarly
to (Haury et al., 2011), we found that average rank ag-
gregation usually performed worse than the other ag-
gregation methods. We also found that average score
aggregation usually resulted, with a few exception, in
the best stability, while stability selection aggregation
was in-between.
We observed, in some cases, a trade-off between
stability and error rate. Previous studies such as
(Saeys et al., 2008) already suggested such a dataset-
dependent trade-off between robustness and classifi-
cation performance. Here, however, we find that the
aggregation method can also play a role in this trade-
off, since in some cases error rate and stability were
differently affected by the different ensemble aggre-
gation methods. This trade-off did not seem to apply
to our artificial data, though: on them, a better er-
ror rate was systematically paired with a better stabil-
ity. This difference could be due to structural differ-
ences with the real data (the latter probably present-
ing much more complex and numerous interactions),
or to a lack of diversity in the artificial data, since the
trade-off is not observed in all datasets. Nonetheless,
we observed that in all cases, the most stable method
without ensemble could be rendered more stable via
ensemble, with or without a trade-off on the classifi-
cation error rate.
As future work, we think it would be interesting
to study or develop more aggregation methods, such
as average weighted rank or score (giving a higher
weight to higher scores or lower ranks). Weighted
(exponential) rank reportedly performed better than
average rank (Haury et al., 2011), so maybe such an
improvement could be obtained by using the same
method on scores. Hybridizationof different base fea-
ture selection methods also seems to be an interesting
area to explore, which will also require some specific
work on the aggregation strategies.
REFERENCES
Abeel, T., Helleputte, T., Van de Peer, Y., Dupont, P., and
Saeys, Y. (2010). Robust biomarker identification
for cancer diagnosis with ensemble feature selection
methods. Bioinformatics, 26(3):392–398.
Dernoncourt, D., Hanczar, B., and Zucker, J.-D. (2014).
Analysis of feature selection stability on high dimen-
sion and small sample data. Computational Statistics
and Data Analysis, 71(0):681 – 693.
Ein-Dor, L., Zuk, O., and Domany, E. (2006). Thousands
of samples are needed to generate a robust gene list
for predicting outcome in cancer. Proceedings of the
National Academy of Sciences, 103(15):5923–5928.
Han, Y., Yang, Y., and Zhou, X. (2013). Co-regularized
ensemble for feature selection. In Proceedings of the
Twenty-Third International Joint Conference on Arti-
ficial Intelligence, IJCAI’13, pages 1380–1386. AAAI
Press.
Han, Y. and Yu, L. (2012). A variance reduction framework
for stable feature selection. Statistical Analysis and
Data Mining, 5(5):428–445.
Haury, A.-C., Gestraud, P., and Vert, J.-P. (2011). The influ-
ence of feature selection methods on accuracy, stabil-
ity and interpretability of molecular signatures. PLoS
ONE, 6(12):e28210.
Jain, A. K. and Chandrasekaran, B. (1982). 39 dimension-
ality and sample size considerations in pattern recog-
nition practice. Handbook of Statistics, 2:835–855.
Kuncheva, L. I. (2007). A stability index for feature selec-
tion. In Devedzic, V., editor, Artificial Intelligence and
Applications, pages 421–427. IASTED/ACTA Press.
Liu, H., Liu, L., and Zhang, H. (2010). Ensemble gene se-
lection by grouping for microarray data classification.
Journal of biomedical informatics, 43:81–87.
Meinshausen, N. and Bhlmann, P. (2010). Stability selec-
tion. Journal of the Royal Statistical Society: Series B
(Statistical Methodology), 72(4):417–473.
Rokach, L. (2010). Ensemble-based classifiers. Artificial
Intelligence Review, 33(1-2):1–39.
Saeys, Y., Abeel, T., and Peer, Y. (2008). Robust feature
selection using ensemble feature selection techniques.
In Daelemans, W., Goethals, B., and Morik, K., edi-
tors, Machine Learning and Knowledge Discovery in
Databases, volume 5212 of Lecture Notes in Com-
puter Science, pages 313–325. Springer Berlin Hei-
delberg.
Simon, R. (2003). Supervised analysis when the number of
candidate features (p) greatly exceeds the number of
cases (n). SIGKDD Explor. Newsl., 5(2):31–36.
Somol, P., Grim, J., and Pudil, P. (2009). Criteria ensembles
in feature selection. In Benediktsson, J. A., Kittler, J.,
and Roli, F., editors, MCS, volume 5519 of Lecture
Notes in Computer Science, pages 304–313. Springer.
Somol, P. and Novoviˇcov´a, J. (2010). Evaluating stability
and comparing output of feature selectors that opti-
mize feature subset cardinality. IEEE Trans. Pattern
Anal. Mach. Intell., 32(11):1921–1939.
Surowiecki, J. (2004). The Wisdom of Crowds: Why the
Many are Smarter Than the Few and how Collective
Wisdom Shapes Business, Economies, Societies, and
Nations. Doubleday.
Wald, R., Khoshgoftaar, T. M., and Dittman, D. J. (2013).
Ensemble gene selection versus single gene selec-
tion: Which is better? In Boonthum-Denecke, C.
and Youngblood, G. M., editors, FLAIRS Conference.
AAAI Press.
Yang, P., Hwa Yang, Y., B. Zhou, B., and Y. Zomaya, A.
(2010). A review of ensemble methods in bioinfor-
matics. Current Bioinformatics, 5(4):296–308.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
330
