
Using Artificial Intelligence to Improve the Evaluation of Human
Blastocyst Morphology
José Celso Rocha
1
, Diogo Lima Bezerra da Silva
1
, João Guilherme Cândido dos Santos
1
,
Lucy Benham Whyte
2,3
, Cristina Hickman
2,4
, Stuart Lavery
2,4
and Marcelo Fábio Gouveia Nogueira
5
1
Laboratório de Matemática Aplicada, FCL, Universidade Estadual Paulista (Unesp),
Av. Dom Antonio 2100, Assis, Brazil
2
Boston Place Clinic, 20 Boston Place, NW16ER, U.K.
3
University of Oxford, U.K.
4
Imperial College London, U.K.
5
Laboratório de Micromanipulação Embrionária, FCL, Unesp, Av. Dom Antonio 2100, Assis, Brazil
Keywords: Artificial Intelligence, Human Embryo, Embryo Classification, Image Digital Processing.
Abstract: The morphology of the human embryo produced by in vitro fertilized (IVF) is historically used as a
predictive marker of gestational success. Although there are several different proposed methods to improve
determination of embryo morphology, currently, all methods rely on a manual, optical and subjective
evaluation done by an embryologist. Given that tiredness, mood and distinct experience could influence the
accuracy of the evaluation, the results found are very different from embryologist to embryologist and from
clinic to clinic. We propose the use of an objective evaluation, with repeatability and automatization, of the
human blastocyst by image processing and the use of Artificial Neural Network (i.e., Artificial Intelligence).
1 INTRODUCTION
Since the establishment of assisted reproduction
techniques (ART) in humans the quality of the
embryos in the blastocyst stage has been shown to
be able to predict the efficacy of the implantation
and the probability of the embryo to generate
pregnancy (della Ragione et al., 2007; Ahlstrm et al.,
2011). The predominant technique currently used to
determine embryo quality is the morphological
analysis by means of optical microscopy; this
method, despite being able to establish predictive
relations with the pregnancy rate, is still subjective
and, in many cases, with limited reproducibility. The
main problem of this method lies in the subjectivity
in the interpretation of the results by the
embryologists, resulting in low interobserver
agreement and intraobserver reproducibility (Arce et
al., 2006; Sundvall et al., 2013; Richardson et al.,
2015)
According to Gardner and Schoolcraft (1999) the
embryo classification is made according to three
parameters: i) stage of expansion and hatching (EE),
classified from 1 to 6, being 1 the embryo without
any inner cavity (blastocoel) meaning that it not
reached the blastocyst stage yet and 6 the blastocyst
fully hatched; ii) quality of the inner cell mass
(ICM) classified from A to C, being A the ICM with
the highest quality and C the worst and; iii) quality
of the trophectoderm (TE), also classified as A to C
and in the same way as ICM. Examples of
blastocysts classified by the Gardner & Schoolcraft
system are shown in Figure 1.
For Gardner and Schoolcraft classification
(1999), the technique used is the morphological
assessment by stereomicroscopy that is non-
invasive, however there are several other methods to
classify blastocysts such as metabolism
measurement (Tejera et al., 2016) and time-lapse
(Tejera, Aparicio-Ruiz and Meseguer, 2017) which
are also non-invasive methods. In addition, there are
techniques such as blastocyst transcriptome analysis
(Kakourou et al., 2013) and chromosomal screening
by array comparative genomic hybridization (aCGH)
(Yang et al., 2012) that are invasive. Invasive
techniques are not appropriate to classify human
embryos as they may jeopardize the integrity of the
embryo and, consequently, decrease the probability
of his implantation.
Rocha J., Bezerra da Silva D., dos Santos J., Whyte L., Hickman C., Lavery S. and Gouveia Nogueira M.
Using Artificial Intelligence to Improve the Evaluation of Human Blastocyst Morphology.
DOI: 10.5220/0006515803540359
In Proceedings of the 9th International Joint Conference on Computational Intelligence (IJCCI 2017), pages 354-359
ISBN: 978-989-758-274-5
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

Figure 1: Illustrative images of human blastocysts
classification. The first number is referred to the presence
and size of the blastocoel as well as the degree of embryo
expansion. The first letter is referred to the quality of the
inner cell mass and (second letter) of the trophectoderm.
(a) 3AA; (b) 3AB; (c) 3BA; (d) 4AA; (e) 4AB; (f) 4BA;
(g) 4CC; (h) 5AA; (i) 5CA. From Van den Abbeel, p.357,
2013.
One of the ways to reduce the subjectivity
involved in that process and to make a more
objective classification is the use of digital image
processing and artificial intelligence (AI) techniques
such as artificial neural networks (ANNs) and
genetic algorithms (GAs). With these methods, it is
possible to obtain a high reproducibility independent
of experience, attention to detail and systematic
approach of the examiner, factors which are
confounding with visual morphological assessment
of human embryos. Such techniques have already
been used to classify mammalian embryos, obtaining
promising results (Matos, Rocha and Nogueira,
2014; Rocha et al., 2016).
The technique of digital image processing
consists of extracting information of size, colour
scale and saturation using mathematical methods
(Gonzalez and Woods, 2007). This technique allows
the extraction of several variables such as
circularity, radius, uniformity, texture, luminosity,
and colour scale from the photos of the blastocysts
(Rocha et al., 2016), which are important for the use
in ANN technique.
Genetic algorithms are algorithms of global
optimization of functions, based on the theory of
natural selection proposed by Charles Darwin. In
this theory, individuals whose phenotype is better
fitted to the environment are more likely to achieve
reproductive success, so they are more likely to
propagate their genes to the next generation. In
addition to this, processes such as recombination,
crossing over and mutation imply in differentiation
between the chromosomes of the offspring and the
parents promoting genetic variation and thus they
evolve increasingly adapting to the environment to
which they are inserted (Lacerda and Carvalho,
1999). In this case the individuals are the ANNs and
the genes are the various parameters that define the
network architecture.
The algorithm works with iterations that are
called generations and for each generation, the
principles of selection, migration, replication and
repopulation are applied to a population of ANN
architectures.
ANNs are based on the biological neuron model,
which can to learn through experience and error. The
main characteristic of a biological neuron is the
ability to receive and interpret stimuli, transmitting
information to nearby neurons (Kovács, 2002). This
learning capacity is achieved through interconnected
neurons in layers that upon receiving a stimulus,
process this information through a weighted value,
called weight, that ends up storing the knowledge of
the ANN. The weights indicate the influence of the
signal at the output of each neuron (Haykin, 2001).
Currently, the greatest difficulty is the determination
of the number of neurons and layers to be used, so
that these are usually obtained through exhaustive
case studies (Jayas, Paliwal and Visen, 2000).
However, with a statistically relevant database
and evolutionary algorithms (like the GAs) the
architecture that best fits the classification problem
can be found more effectively (Schaffer, Whitley
and Eshelman, 1992).
Tools such as time-lapse monitoring - present in
some equipment as EmbryoScope
®
- have been used
for observation and data retrieval from human
embryos, without limiting the number of
observations made (i.e., images obtained). By this
technology, coupled with an appropriate software, a
video is produced and it reports the embryonic
development during the in vitro culture period.
Through this, much information is provided on the
whole process of morphological transformations
occurring in the embryo, such as kinetics and
asymmetry of cleavages (Kovacs, 2014).
The aim of the present work is to use the time-
lapse monitoring to extract images of human
blastocysts at a specific moment post-insemination
and submit these images to the digital processing
techniques to obtain mathematical variables
representatives of the embryos. After this step and
using AI techniques, we intend to obtain, through a
computer software, an automatized classification of
human blastocysts images as already developed for
the bovine species (Rocha et al., 2016, 2017).
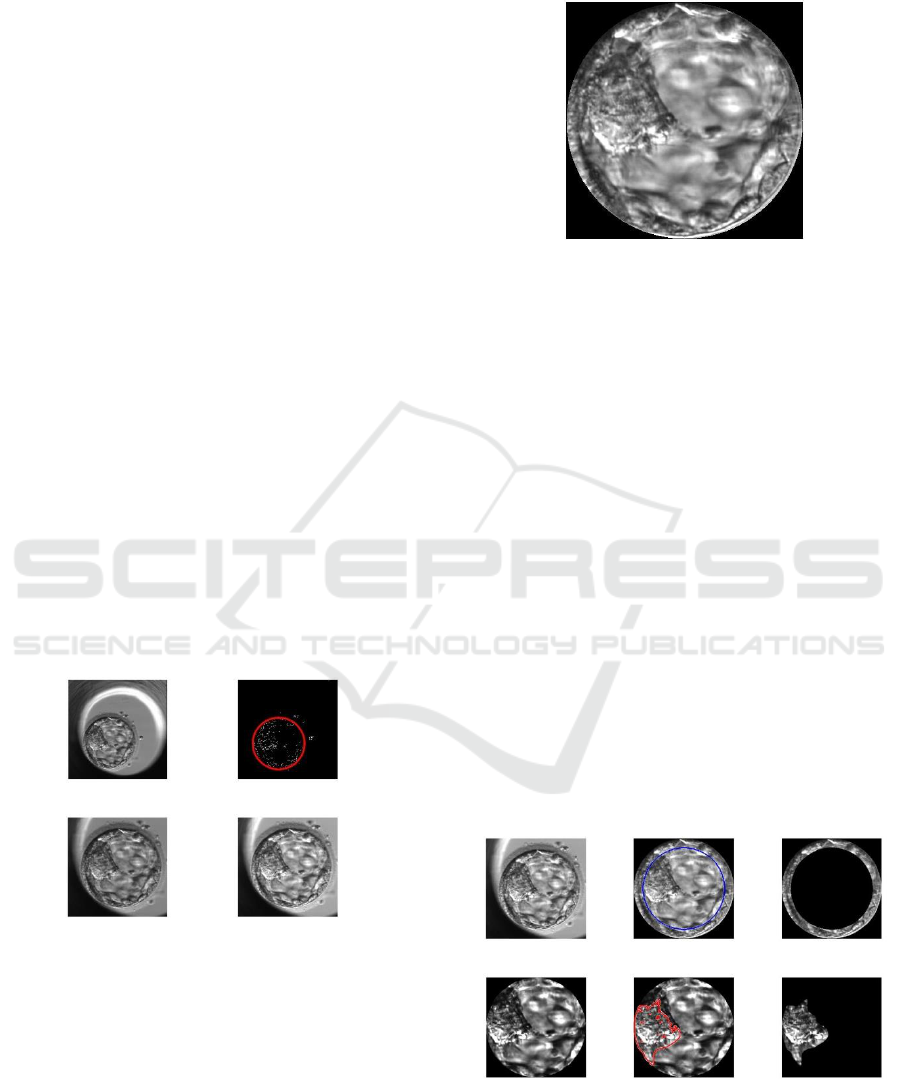
The images of human blastocysts used in the
digital processing, as well as their classification, that
will be used for the AI technique, were provided by
the London-based Boston Place Clinic, which is our
partner in the development of this work.
2 METHODOLOGY
2.1 Digital Image Processing
Images of human blastocysts, obtained through
EmbryoScope
®
by the Boston Place Clinic, were
standardized to have the same resolution and
illumination characteristics. The proposed algorithm
automatically imports the image into the MatLab
®
software environment, and standardizes the image
by converting the image into grayscale, adjusting the
resolution and the aspect ratio. Conversion to
grayscale allows for avoidance of the variation due
to colour, thus all images are converted to 8-bit gray
scale. This process provides a higher speed in the
processing of the next steps, as it decreases the
spectral dimension of the image. To solve the
problem of the different illuminations of the images,
a histogram adjustment was made. In the image, 1%
of all information was saturated between light and
dark pixels, increasing the contrast of the image and,
thus, facilitating the next step of segmentation.
Figure 2 shows the standardization of a human
blastocyst.
Figure 2: Human blastocyst standardization.
After standardization, the blastocyst was isolated
from the rest of the image (i.e., background) before
the extraction of the variables. This process consists
in altering the image gradient so that the limits of the
blastocyst become more evident. For this step, the
Hough's Transform function was used (Atherton and
Kerbyson, 1999), which delineates the
circumference that best characterizes the blastocyst.
An example of the isolated blastocyst is shown in
Figure 3.
Figure 3: Isolated human blastocyst by Hough’s
Transform.
The complete image processing is performed
using several algorithms that act individually as
Gray Level Co-Occurrence Matrix (GLCM)
(Haralick, Shanmugam and Dinstein, 1973) for
texture analysis, the Watershed Transform, which
seeks to segment the image (Beucher, 1992) in
addition to the Gabor filter that differentiates the
various textures of the image through the
characterization of a signal simultaneously in the
time domain and in the domain of the spatial
frequencies (Marmol, 2011). After the application of
these techniques, the TE and the ICM were
separately identified, whilst isolating the blastocyst
completely. The complete processing of an
illustrative image of the human blastocyst is
demonstrated in Figure 4.
Following the process of image segmentation, a
numerical vector is derived that will represent the
extracted characteristics of the images. This vector
will be used as input variable for the ANN, thus
making the image-derived information proper for
use in computational techniques.
Figure 4: An illustrative sequence of the complete
processing of the human blastocyst image. In the upper
row (left) it is the original image without processing. In
the right column, it is shown the trophectoderm mask
(upper) and the inner cell mass (lower) after segmentation.

2.2 Artificial Intelligence
After obtaining the variables, that identify the
human embryo and that will be used for the GA
technique, a population of individuals will be
constructed – which represent several architectures
of the ANNs in their chromosomes. The
chromosomes will be randomly generated forming
an initial population of individuals. Each population
will contain from 100 to 200 individuals. Each
chromosome, which will represent a specific ANN,
will contain in its genes the maximum and minimum
number of neurons per layer, the number of layers to
be used, the learning rate, the transfer functions to be
used (logsig, purelin, tansig, hardlim, tribas, radbas
or satlin) and the learning functions (trainrp,
trainscg, traincgb, traincgf, traincgp, traingdm or
traingd) (Beale, Hagan and Demuth, 2017). The
entire process will be developed in the MatLab
®
environment (MATLAB 2017a, The MathWorks
Inc., Natick, MA) that has tools for creating and
modelling ANNs.
After the generation of the ANNs (individuals),
the entire population will be trained, validated, and
tested using the blastocyst images database, which
will be divided into training (from 50% to 70% of
the data), validation and test (can be 15% to 25%
each) sets.
For the following generations, 20% of the
selected individuals will be kept as the fittest, 60%
will be composed by the recombination and
mutation of the individuals of the previous
population and the remaining 20% will come from
the migration. The number of generations will be a
maximum of 1000. The most fitted individuals will
be chosen from the smallest error of the test set
when applying the ANN technique.
It is intended that at the end of the iterations,
previously established, the software will present an
optimized ANN architecture that classifies the
human embryos in a less subjective way and with
greater reproducibility and assertiveness. Of course,
the whole processing will be in an automatized way
(i.e., without human intervention unless the upload
of the original image).
3 DISCUSSION
Currently, we have observed that the human
blastocyst images, in terms of digital processing, is
quite different from the mouse and bovine
blastocysts already studied in previous research
(Van Soom et al., 2003; Matos, Rocha and
Nogueira, 2014; Rocha et al., 2016, 2017)
Differently from murine and bovine blastocysts,
which present well defined ICM and blastocoel at
the time of implantation, human blastocysts have a
huge ICM variation in terms of shape that,
consequently, decreases the accuracy of ICM
masking by digital processing (Figure 5).
Figure 5: Illustrative images of mouse, bovine and human
blastocysts (from left to right) to show the differences
found on the inner cell mass shape mainly on the human
embryos. Asterisk (*) marks the inner cell mass on each
image.
This fact can difficult in the determining the
mathematical variables that characterize the human
embryo and, consequently, the ANN inputs. Those
inputs, if not properly extracted from the image, will
not be representative of the blastocyst and thus the
ANN will be wrongly trained.
The next step is to enhance the way to obtain the
fittest mask of the isolated ICM since the mask of
isolated TE already seems fitted. In this way, it is
essential to choose carefully what frame coming
from the time-lapse record will be used on the image
processing, since the same embryo in a short time
frame could be registered with different images by
the equipment.
4 CONCLUSIONS
Although in its early steps of development, the
automatized, reproducible, and objective evaluation
of human blastocysts by AI, is a promising tool to
improve the way that in the future the embryologist
could choose which embryo should be transferred to
the patient. Since this proposed method is based on a
long previous study with mouse and bovine
blastocysts, to adapt the knowledge previously
obtained to the human scope would be not a
hindrance.

ACKNOWLEDGEMENTS
Grants supporting the authors’ research
#2012/50533-2, #2013/05083-1, #2006/06491-2,
#2011/06179-7 and 2012/20110-2 from São Paulo
Research Foundation (FAPESP). We also thank
Agência UNESP de Inovação (AUIN) for processing
the national and international patents of the
invention.
REFERENCES
Ahlstrm, A. et al. (2011) ‘Trophectoderm morphology: An
important parameter for predicting live birth after
single blastocyst transfer’, Human Reproduction,
26(12), pp. 3289–3296. doi: 10.1093/humrep/der325.
Arce, J. et al. (2006) ‘Interobserver agreement and
intraobserver reproducibility of embryo quality
assessments’, 21(8), pp. 2141–2148. doi: 10.1093/
humrep/del106.
Atherton, T. J. and Kerbyson, D. J. (1999) ‘Size invariant
circle detection’, Image Vision Computing, 17, pp.
795–803.
Beale, M. H., Hagan, M. T. and Demuth, H. B. (2017)
Neural Network ToolboxTM User’s Guide.
Beucher, S. (1992) ‘The watershed transformation applied
to image segmentation’, Scanning Microscopy
Supplement, 6, pp. 299–314.
Gardner D. K., Schoolcraft W. B. (1999) In vitro culture
of human blastocyst. In: Janson R, Mortimer D, eds.
Towards Reproductive Certainty: Infertility and
Genetics Beyond. Carnforth: Parthenon Press. p.378-
88.
Gonzalez, R. C. and Woods, R. E. (2007) Digital Image
Processing. 3rd edn. Pearson Prentice Hall.
Haralick, R. M., Shanmugam, K. and Dinstein, I. (1973)
‘Textural Features for Image Classification’, IEEE
Transactions on Systems, Man and Cybernetics, 3, pp.
610–621.
Haykin, S. (2001) Redes Neurais: princípios e prática.
2nd edn. Porto Alegre: Bookman.
Jayas, D. S., Paliwal, J. and Visen, N. S. (2000) ‘Review
Paper (AE—Automation and Emerging
Technologies)’, Journal of Agricultural Engineering
Research, 77(2), pp. 119–128. doi: 10.1006/
jaer.2000.0559.
Kakourou, G. et al. (2013) ‘Investigation of gene
expression pro fi les before and after embryonic
genome activation and assessment of functional
pathways at the human metaphase II oocyte’, Fertil
Steril. doi: 10.1016/j.fertnstert.2012.10.036.
Kovacs, P. (2014) ‘Embryo selection: the role of time-
lapse monitoring’, Reproductive Biology and
Endocrinology, 12, p. 124.
Kovács, Z. L. (2002) Redes Neurais Artificiais. Editora
Livraria da Fisica.
Lacerda, E. G. M. De and Carvalho, A. C. P. L. F. de
(1999) Introdução aos Algoritmos Genéticos.
Marmol, U. (2011) ‘Use of Gabor filters for texture
classification of airborne images and LIDAR data’,
Archiwum Fotogrametrii, Kartografii i Teledetekcji,
22, pp. 325–336. Available at: http://ptfit.sgp.
geodezja.org.pl/wydawnictwa/krakow2011/APCRS
vol. 22 pp. 325-336.pdf5-336.pdf.
Matos, F. D., Rocha, J. C. and Nogueira, M. F. G. (2014)
‘A method using artificial neural networks to
morphologically assess mouse blastocyst quality’,
Journal of Animal Science and Technology, 56.
MATLAB R2017a. Natick, MA: MathWorks, 2017.
Computer software.
della Ragione, T. et al. (2007) ‘Developmental stage on
day-5 and fragmentation rate on day-3 can influence
the implantation potential of top-quality blastocysts in
IVF cycles with single embryo transfer.’,
Reproductive biology and endocrinology : RB&E, 5, p.
2. doi: 10.1186/1477-7827-5-2.
Richardson, A. et al. (2015) ‘A clinically useful simplified
blastocyst grading system’, Reproductive BioMedicine
Online, 31, pp. 523–530.
Rocha, J. C. et al. (2016) ‘Methods for assessing the
quality of mammalian embryos: How far we are from
the gold standard?’, JBRA Assisted Reproduction,
20(3), pp. 150–158.
Rocha, J. C. et al. (2017) ‘A method based on artificial
intelligence to fully automatize the evaluation of
bovine blastocyst images’, Scientific Reports,
7(Article number: 7659).
Schaffer, J. D., Whitley, D. and Eshelman, L. J. (1992)
‘Combinations of genetic algorithms and neural
networks: a survey of the state of the art’,
[Proceedings] COGANN-92: International Workshop
on Combinations of Genetic Algorithms and Neural
Networks, pp. 1–37. doi: 10.1109/
COGANN.1992.273950.
Van Den Abbeel. et al. (2013) ‘Association between
blastocyst morphology and outcome of single-
blastocyst transfer’, Reproductive biomedicine online,
v. 27, n. 4, p. 353-361.
Van Soom, A. et al. (2003) ‘Assessment of mammalian
embryo quality: what can we learn from embryo
morphology?’, Reproductive biomedicine online.
Reproductive Healthcare Ltd, Duck End Farm, Dry
Drayton, Cambridge CB23 8DB, UK, 7(6), pp. 664–
70. doi: 10.1016/S1472-6483(10)62089-5.
Sundvall, L. et al. (2013) ‘Inter- and intra-observer
variability of time-lapse annotations’, 28(12), pp.
3215–3221. doi: 10.1093/humrep/det366.
Tejera, A. et al. (2016) ‘Combination of metabolism
measurement and a time-lapse system provides an
embryo selection method based on oxygen uptake and
chronology of cytokinesis timing’, Fertility and
sterility, 106, pp. 119–126. doi: 10.1016/
j.fertnstert.2016.03.019.
Tejera, L., Aparicio-Ruiz, B. and Meseguer, M. (2017)
‘The use of morphokinetic as a predictor of
implantation’, MINERVA GINECOLOGICA. doi:
10.23736/S0026-4784.17.04098-9.

Yang, Z. et al. (2012) ‘Selection of single blastocysts for
fresh transfer via standard morphology assessment
alone and with array CGH for good prognosis IVF
patients: results from a randomized pilot study’,
Molecular Cytogenetics, 5(1), p. 24. doi:
10.1186/1755-8166-5-24.
