
An Automated Method for Generating Training Sets for Deep Learning
based Image Registration
Masato Ito and Fumihiko Ino
Graduate School of Information Science and Technology, Osaka University,
1-5 Yamadaoka, Suita, Osaka 565-0871, Japan
Keywords:
Image Registration, Nonrigid Registration, Deep Learning, Training Data.
Abstract:
In this paper, we propose an automated method for generating training sets required for realizing deep learning
based image registration. The proposed method minimizes effort for supervised learning by automatically
generating thousands of training sets from a small number of seed sets, i.e., tens of deformation vector fields
obtained with a conventional registration method. To automate this procedure, we solve an inverse problem
instead of a direct problem; we produce a floating image by applying a deformation vector field Φ to a reference
image and let the inverse vector of Φ be the ground truth for these images. In experiments, the proposed
method took 33 minutes to produce 169,890 training sets from approximately 670,000 2-D magnetic resonance
(MR) images and 30 seed sets. We further trained GoogLeNet with these training sets and performed holdout
validation to compare the proposed method with the conventional registration method in terms of recall and
precision. As a result, the proposed method increased recall and precision from 50% to 80%, demonstrating
the impact of deep learning for image registration problems.
1 INTRODUCTION
Image registration (Hajnal et al., 2001) is a tech-
nique for defining a geometric relationship between
each point in two different images: a reference image
and a floating image. This technique eliminates ge-
ometric gaps between two clinical images, which are
caused due to various factors: different cycles of pa-
tient’s respiration, different modalities, different pa-
tients, and so on. The aligned images are useful for
medical doctors to know the exact shape and location
of the tumor. In particular, nonrigid registration algo-
rithms are required to align deformable organs such
as the brain (Rohlfing and Maurer, 2003), liver (Ino
et al., 2005a), lung (Ino et al., 2005b), and etc.
Many registration algorithms (Rueckert et al.,
1999; Klein et al., 2009) define a similarity measure
between two images, which is then used as a cost
function to be optimized. Because this optimization-
based approach involves a large amount of compu-
tation, many parallel machines such as clusters (Ino
et al., 2005a) and graphics processing units (GPUs)
(Ikeda et al., 2014) have been deployed for acceler-
ated optimization. Although this optimization process
was automated in previous systems (Rueckert et al.,
1999; Ino et al., 2005a; Ikeda et al., 2014), the align-
ment process results in an alignment failure if the op-
timizer falls into a local solution. Therefore, we need
a better optimizer to realize accurate and robust regis-
tration systems.
In contrast to these conventional algorithms, deep
learning (Goodfellow et al., 2016) has emerged as
a new machine learning technique in various fields
such as image classification, speech recognition, rec-
ommendation engine, and so on. This technique in-
creases classification accuracy with neural networks
(NNs), which have many deep layers in the network
topology. These deep layers allow classification sys-
tems to learn complicated and abstract features hidden
in the input data, which seems to be hard for conven-
tional techniques to learn. However, deep learning
requires enormous training sets to increase the classi-
fication accuracy. Therefore, some training sets such
as ImageNet (Deng et al., 2009) are freely available
for specific applications (i.e., image classification),
where deep learning has been widely used. However,
these useful datasets are not available for unexplored
applications, where deep learning has not been widely
used yet. With respect to image registration, train-
ing NNs requires ground truth of deformation vec-
tor fields, which give voxel correspondence between
reference and floating images. Given a pair of three-
dimensional (3-D) images of n
3
voxels, obtaining an
140
Ito, M. and Ino, F.
An Automated Method for Generating Training Sets for Deep Learning based Image Registration.
DOI: 10.5220/0006634501400147
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 2: BIOIMAGING, pages 140-147
ISBN: 978-989-758-278-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

n
3
deformation vectors, each for a voxel in the float-
ing image, is hard to generate by hand. Thus, the lack
of training sets is an important issue when applying
deep learning to image registration problems.
As for image classification, training sets can be
easily generated from a small number of seed sets
by adding subtle changes. For example, many re-
searchers (Krizhevsky et al., 2012; Howard, 2013;
Szegedy et al., 2015) created training sets by adding
noise, changing image resolution, and altering aspect
ratio. However, these approaches cannot be used for
image registration problems because the approaches
compromise the correctness of deformation vectors;
that is, the number of correct deformation vector
fields cannot be automatically increased. Another ap-
proach is to use datasets correctly aligned with con-
ventional registration methods. However, this ap-
proach fails to improve the registration accuracy be-
cause NNs trained with such datasets never achieve
higher accuracy than that achieved with conventional
algorithms. Therefore, automating training set gener-
ation is a critical issue for applying deep learning to
image registration problems.
In this paper, we propose a method for minimiz-
ing effort required for obtaining a large number of
training sets for image registration. The key idea
for automating training set generation is to consider
inverse problems instead of direct problems. The
proposed method generates O(N) deformation vector
fields (i.e., ground truth data) from N reference im-
ages and M deformation vector fields (i.e., seed data);
the seed data here are obtained with a conventional
registration method and are validatedtheir correctness
by hand. Such realistic deformation vector fields are
useful for variating deformed images with seed sets;
we apply each vector field to different reference im-
ages. In other words, the reference images exactly
match with those transformed from the deformed im-
ages with inverse vectors of the deformation fields.
Therefore, the proposed method stores the inverse in-
formation as the ground truth for the reference and
transformed images. Furthermore, we realize deep
learning based registration by solving a classification
problem; voxels are classified according to their de-
formation direction.
The remainder of the paper is organized as fol-
lows. Section 2 introduces related work on regis-
tration with deep learning. Section 3 presents an
overview of conventional registration methods that
solve a registration problem into an optimization
problem. Section 4 describes our proposed method.
Section 5 shows several experimental results obtained
with the latest GPU. Finally, Section 6 concludes the
paper with future work.
2 RELATED WORK
Ghosal et al. (Ghosal and Ray, 2017) used a fully
convolutional NN (CNN) to increase the accuracy of
image registration. Their registration algorithm de-
ployed sum of squared difference as the cost func-
tion to be optimized. Deep learning techniques such
as fully CNNs and backpropagation were used to ex-
tract features hidden in reference images. However,
these techniques were used to modify reference im-
ages without taking advantage of learning.
Wu et al. (Wu et al., 2013) realized unsupervised
deep feature learning to improve the accuracy of reg-
istration methods such as Demons (Vercauteren et al.,
2009) and HAMMER (Shen and Davatzikos, 2002).
They detected key image features by compressing im-
ages with a stacked autoencoder that replaced a prin-
cipal component analyzer. Thus, deep learning tech-
niques were used for feature extraction of images; the
optimization framework relied on conventional tech-
niques rather than deep learning techniques. By con-
trast, we tackle to integrate deep learning techniques
into the optimization framework.
Cheng et al. (Cheng et al., 2016) developed a NN
classifier capable of evaluating the similarity measure
for multimodal images. Their registration algorithm
was based on a conventional optimization framework.
Furthermore, training sets were fully collected by
hand, and thus, initial effort was critical when uti-
lizing their approach for image registration problems.
The proposed method reduces manual intervention by
automating the major part of training set generation;
seed sets must be investigated by hand.
3 IMAGE REGISTRATION BASED
ON INFORMATION THEORY
Let r and f be a reference image and a floating im-
age, respectively. Let S(r, f) be a similarity mea-
sure between images r and f. An image registra-
tion problem then can be regarded as an optimiza-
tion problem that finds a geometrical transformation
T : (x,y,z) 7→ (x
′
,y
′
,z
′
) such that S(r, T( f)) is maxi-
mized. Transformation T here maps a voxel (x,y, z) in
image f to its corresponding voxel (x
′
,y
′
,z
′
) in image
r.
As such transformation T, previous methods
(Rueckert et al., 1999; Ikeda et al., 2014) used a B-
spline deformation model (Rueckert et al., 1999; Lee
et al., 1997) to realize free-form deformation with
less amount of computation. As shown in Fig. 1,
this model represents object deformation by moving
control points placed on the image domain. These
An Automated Method for Generating Training Sets for Deep Learning based Image Registration
141
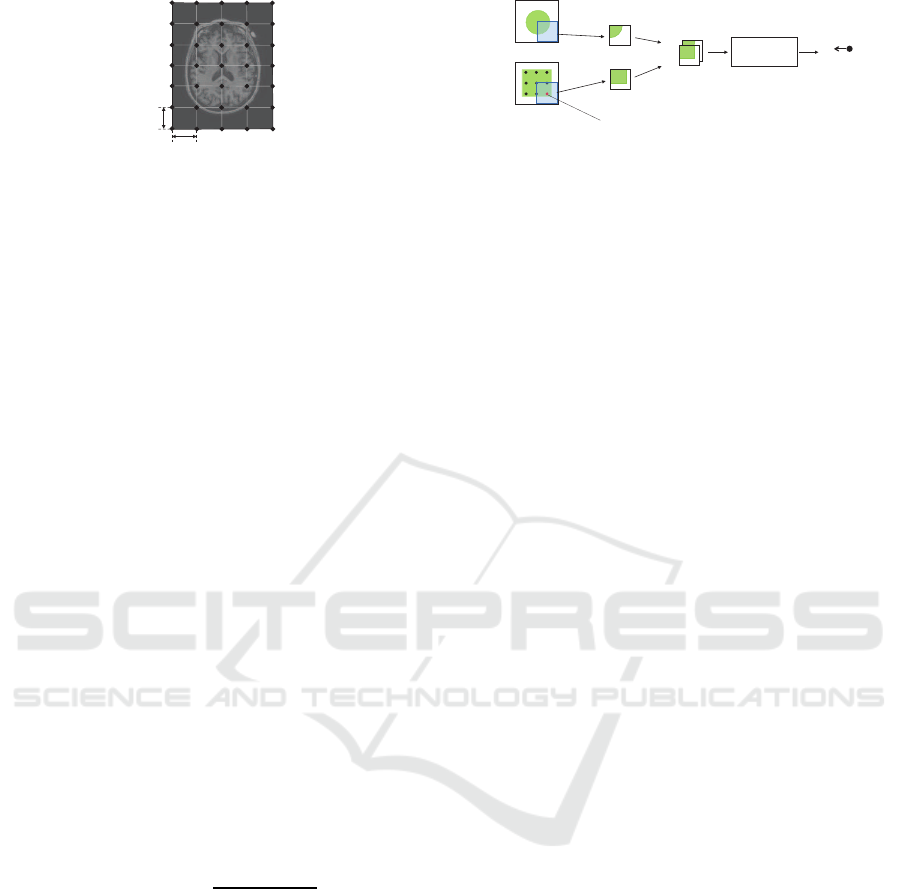
G
G
Figure 1: A control mesh required for B-spline deforma-
tion. Control points are initially placed with intervals of δ
to realize free-form deformation with less amount of com-
putation.
control points are initially placed with common in-
tervals of δ and organized in a hierarchy. Global and
coarse-grained deformation can be realized with large
δ, whereas local and fine-grained deformation can be
realized with small δ. In the following discussion, let
Φ be a control mesh, or a set of control points that
represents the deformation vector field.
Based on this light-weight deformation model, the
previous methods solve a registration problem into an
optimization problem. In more detail, the methods
find a transformation T
opt
that minimizes a cost func-
tion C
T
opt
= argmin
T
C(r,T( f)). (1)
A gradient descent method is typically used for opti-
mization of the cost function C (Φ) given by
C(Φ) = −S
NMI
(Φ), (2)
where S
NMI
represents normalized mutual informa-
tion deployed as the similarity measure between
two images. This information theory based mea-
sure is useful for alignment of multimodal images
(Studholme et al., 1999). The normalized mutual in-
formation between images r and f is given by
S
NMI
(r, f) =
H(r) + H( f)
H(r, f)
, (3)
where H(r) and H( f) are entropies of images r and f,
respectively, and H(r, f) is the joint entropy of r and
f.
During optimization, the gradient descent method
iteratively computes ∂C /∂φ, the partial differential of
the cost function, for all control points φ ∈ Φ. Accord-
ing to this partial differential, φ moves to a direction
such that C(Φ) is minimized.
Moreover, the previous methods organize the im-
ages with a hierarchy of L levels to accelerate reg-
istration process; with this hierarchical organization,
the images are aligned from coarse to fine levels. As
we mentioned before, the control mesh is organized
into the hierarchy accordingly.
Reference image r
Floating image f
Subimage r’
Subimage f’
Control point
I
Deformation
direction
2-channel
image
Trained
GoogLeNet
Figure 2: An overview of the proposed method. Given a
pair of subimages with a control point, the method estimates
the deformation direction for the control point.
4 PROPOSED METHOD
The proposed method realizes image registration in
the following phases:
1. Seed set generation. We use the conventional reg-
istration method (Ikeda et al., 2014) to generate
seed sets. The seed sets here are realistic defor-
mation vector fields generated from successfully-
aligned cases.
2. Training set generation. The proposed method au-
tomatically variates training sets using the seed
sets.
3. CNN training. A CNN, which gives the deforma-
tion vector fields for real-world data, are trained
with the generated training sets.
4. Inference. The trained CNN is used to produce the
deformation vector fields for unknown real-world
data.
At the second phase, the proposed method auto-
matically generates training sets without manual in-
tervention. Therefore, a large number of training
sets can be easily generated from a small number of
seed sets. However, as shown in the first phase, the
proposed method relies on the previous registration
method, which means that users have to visually con-
firm whether the seed sets are successfully aligned or
not. In this sense, the proposed method is not a fully
automated method but we think that the effort for col-
lecting a limited number of seed sets are acceptable in
practical cases.
In the following, we first describe the details on
the last phase, i.e., how registration problems are
solved into optimization problems. We then sum-
marize on the first phase, i.e., how the conventional
method generates the deformation vector fields. Fi-
nally, we present the second phase, i.e., how the pro-
posed method generates training sets from seed sets.
BIOIMAGING 2018 - 5th International Conference on Bioimaging
142

Algorithm 1 Training set generation algorithm
Input: (1) set of N reference images, R = { r
1
,r
2
,..., r
N
}, (2) set of control meshes, Φ = {Φ
m,h
| 1 ≤ m ≤ M,1 ≤
h ≤ L}, where M is the number of seeds, L is the number of hierarchies, and Φ
m,h
represents the mesh of
control points obtained for the m-th seed at the h-th hierarchy, and (3) diameter D of subimages.
Output: set of training data, S = {(r
′
, f
′
,l)}, where r
′
and f
′
are reference and floating subimages, respectively,
and l is the label representing the deformation direction.
1: S ⇐ empty set;
2: foreach r ∈ R do
3: m ⇐ random number between 1 and M;
4: foreach Φ ∈ {Φ
m,1
,Φ
m,2
,... ,Φ
m,L
} do
5: f ⇐ image deformed from r with Φ;
6: foreach φ ∈ Φ do
7: r
′
⇐ subimage extracted from r with distance D of φ;
8: f
′
⇐ subimage extracted from f with distance D of φ;
9: if neither r
′
nor f
′
is blank then
10: l ⇐ label computed with inverse vector of φ;
11: Add (r
′
, f
′
,l) to S ;
12: end if
13: end for
14: end for
15: end for
4.1 Optimization Scheme
The proposed method utilizes deep learning to esti-
mate deformation direction for every control point.
Therefore, the proposed method replaces gradient
computation of the conventional registration method
with inference of deformation direction. In general,
control points in 3-D images takes one of the six di-
rections: ±x, ±y and ±z. Consequently, this estima-
tion problem can be regarded as a classification prob-
lem with six direction classes.
Figure 2 shows the estimation flow of the pro-
posed method. The inputs of the proposed method
are (1) a control point φ, (2) a subimage f
′
containing
the control point φ and its neighboring voxels within
diameter D, and (3) a subimage r
′
covering the same
coordinates of f
′
. Given these inputs, the proposed
method outputs a label l, which indicates the defor-
mation direction for φ. The diameter D must be deter-
mined experimentally such that the longest deforma-
tion is covered within the extracted subimage.
Notice here that the proposed method requires two
input images, r and f, to perform estimation. On
the other hand, NNs for object classification problems
deal with a single image to be classified. To eliminate
this gap on the number of input images, we store r
and f as a single image of two channels.
As for a NN, the proposed method adopts
GoogLeNet (Szegedy et al., 2015), which achieved
a high classification rate for a classification problem
of 1000 object classes. This high rate comes from
its network topology, which consists of full inception
modules that have convolution layers and pooling lay-
ers.
4.2 Seed Data Generation
The proposed method deploys a conventionalregistra-
tion method (Ikeda et al., 2014) to obtain M seed sets
that contain realistic deformation vector fields. Be-
cause the conventional method can fail to align im-
ages, the proposed method requires users to manually
select successfully-aligned images. In more detail,
clinical images of the same patient (but acquired at
different times) are aligned and visually investigated
the correctness of aligned images.
Note here that this investigation collects realistic
deformation vector fields as seed sets, and thereby
the correctness of aligned images can be roughly con-
firmed; we do not require voxel-to-voxel confirma-
tion because the deformation vector fields are used as
seeds for different reference and floating images; the
original reference and floating images, which gener-
ated the deformation vector fields, are not included in
the seeds sets. Hence, the effort for seed investiga-
tion is proportional to M rather than the number n
3
of voxels, meaning that the initial effort is not criti-
cal. In Section 4.3, we explain how the ground truth
data, i.e., training sets, are generated with the pro-
posed method.
Another possible approach for seed set generation
is to use a randomized algorithm that arbitrarily gen-
An Automated Method for Generating Training Sets for Deep Learning based Image Registration
143

Reference image r
+
f = T(r)
rƍ
v
Iƍ
Deformation
vector field )
(a)
Floating image f
+
r = T(f )
Iƍ
ív
rƍ
Inverse of )
(b)
Figure 3: Principles of training set generation that solves
an inverse problem. (a) A floating image is generated from
a reference image with a realistic deformation vector field
obtained with a conventional registration method. (b) The
inverse vector of the deformation field is the ground truth
for the reference and floating images.
erates deformation vector fields. However, this ap-
proach is not realistic because the generated defor-
mation vector field can include implausible control
points. For example, a control point of the B-spline
deformation model depends on its four neighboring
control points. In this case, handling these neighbor-
ing control points at a time is more convincing to in-
crease the accuracy of deformation direction of con-
trol points.
4.3 Training Set Generation
The proposed method outputs a set S of training data
from several inputs: (1) N reference images, M (≪ N)
seed sets of deformation vector fields Φ, and the di-
ameter D of subimages. Each training data here con-
sists of a tuple (r
′
, f
′
,l), where r
′
and f
′
represent the
reference and floating subimages, respectively, and l
is the label for a control point φ that indicates one of
the six deformation directions. Algorithm 1 shows
our training set generation algorithm.
The key idea for obtaining the ground truth of the
deformation vector is to generate the floating subim-
age f
′
from the reference subimage r
′
with a deforma-
tion vector obtained with the conventional registration
method (Fig. 3). In more detail, we generate a float-
ing subimage f
′
by applying the deformation vector v
to the reference subimage r
′
. The inverse vector −v
is then the ground truth for a pair of r
′
and f
′
because
applying −v to f
′
produces r
′
. This procedure can be
iterated with different reference images. In this way,
the generated training sets can be easily grown with
ground truth information.
Each reference image r is used only once to avoid
generating similar data; we randomly select a seed set
Table 1: Execution parameters.
Hierarchical level h 1 2
Interval δ (mm) 16 8
Pixel size (mm) 2.4× 2 1.2× 1
Image resolution (pixels) 85× 128 170 × 256
Subimage resolution 32× 32 32× 32
D× D (pixels)
and apply the selected seed (i.e., the deformation vec-
tor field) to reference images. Consequently, N float-
ing images are generated from N reference images
and M deformation vector fields; in this way, we vari-
ate training sets assuming that M ≪ N; furthermore,
each floating image produces a control mesh con-
taining deformation vectors, each for a control point.
We also exclude inappropriate images that have many
background (i.e., blank) voxels. Such blank images
should be excluded from training sets because blank
voxels make it hard to extract image features during
learning.
Another seed generation approach is to use all
inputs and outputs of the conventional registration
method; the pair of floating and reference images, and
their deformation vector field. However, these vectors
can include wrong vectors even if the aligned results
look correct. Consequently, the learning quality can
be degraded due to wrong labels included in training
sets. We avoid this degradation by adapting the in-
puts to the outputs of the conventional method, i.e.,
the deformation vector field.
5 EXPERIMENTAL RESULTS
To evaluate the proposed method, we generated train-
ing sets from seed sets and trained the GoogLeNet
(Szegedy et al., 2015) with the generated training
sets. We then used the trained GoogLeNet to esti-
mate deformation directions for validation data. The
proposed method was compared with a conventional
registration method (Ikeda et al., 2014) with respect
to precision and recall.
Our experimental machine, running on Win-
dows 10 OS, had an Intel Core i7 5930K 3.5 GHz
CPU, an NVIDIA GeForce GTX 1080 GPU, and
an Intel 535 series SATA solid state drive (SSD).
As an underlying deep learning framework, we de-
ployed Chainer 1.23.0 (Preferred Networks, inc.,
2017), cuDNN 5.1 (NVIDIA Corporation, 2017b),
and CUDA 8.0 (NVIDIA Corporation, 2017a).
As for seed and validation sets, we randomly se-
lected 188 datasets from Alzheimer’s Disease Neu-
roimaging Initiative (ADNI) dataset (Wyman et al.,
BIOIMAGING 2018 - 5th International Conference on Bioimaging
144

Table 2: Distribution of labels associated with generated
training sets. The learning sets and validation sets were dis-
joint.
Label Right Up Left Down
Learning sets 35,361 33,407 30,104 31,724
Validation sets 10,797 9,987 8,709 9,801
2013), which consists of 3-D MR brain images. Due
to the limitation of our current implementation, we
used 2-D slices of these 3-D images, but 3-D images
can be easily supported using additional two labels
(front and back). The resolution of each slice was
170× 256 pixels.
The GoogLeNet assumes that the image resolution
is set to 224× 224 pixels, so that we adapted subim-
age resolution from D× D pixels to 224× 224 pixels;
this operation was done with the OpenCV library. We
also normalized pixel values within the range [0,1] us-
ing a batch normalization method (Ioffe and Szegedy,
2015) because ADNI datasets had different contrasts.
5.1 Training Set Generation
We first generated M = 30 seed sets using the conven-
tional method (Ikeda et al., 2014) with execution pa-
rameters shown in Table 1. This registration process
was carried out with two layers from coarse to fine
levels. We then automatically extracted subimages r
′
and f
′
from reference image r and floating image f,
respectively. The diameter D of subimages was exper-
imentally set to 32 pixels such that the diameter was
long sufficient to cover the coordinates of the trans-
formed control point. We used the same diameter for
all image layers.
Training set generation was completed within 33
minutes on the CPU. We generated 169,890 training
sets from M = 30 seed sets (Fig. 4). Note that 88%
of execution time was spent for input/output access
to the SSD storage. We then split the training sets
into two disjoint groups to perform holdout validation
described later; a quarter was for validation and the
remaining was for learning. Table 2 shows the dis-
tribution of the labels associated with training sets.
Thus, the learning sets and validation sets were dis-
joint, which is necessary to evaluate the generaliza-
tion capacity of CNNs.
5.2 Validation of Estimated Results
We trained the GoogLeNet with the generated train-
ing sets for 50 epochs. This training phase took ap-
proximately 16.3 hours on the deployed GPU. After
that, we estimated the deformation directions for the
validation sets using the trained network. Similarly,
r
′
f
′
(a)
r
′
f
′
(b)
r
′
f
′
(c)
r
′
f
′
(d)
Figure 4: Examples of generated training sets associated
with labels (a) right, (b) up, (c) left, and (d) down.
0
0.2
0.4
0.6
0.8
1
Right Up Left Down
Precision
Label
Proposed method Conventional method
(a)
0
0.2
0.4
0.6
0.8
1
Right Up Left Down
Recall
Label
Proposed method Conventional method
(b)
Figure 5: Inference results for the proposed and conven-
tional methods. (a) precision and (b) recall.
we also executed the conventional method to estimate
the deformation directions for the validation sets.
Figure 5 shows precision and recall obtained with
the validation sets. For both metrics, the proposed
method achieved more than 80% accuracy, whereas
the conventional method reached at most 50% accu-
racy. Thus, the proposed method was more accurate
than the conventional method in terms of the defor-
mation direction of control points. Consequently, we
An Automated Method for Generating Training Sets for Deep Learning based Image Registration
145

0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
0 5 10 15 20 25
Accuracy
# of epochs
Training accuracy Testing accuracy
(a)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
0 5 10 15 20 25
Accuracy
# of epochs
Training accuracy Testing accuracy
(b)
Figure 6: Success rate with different numbers of training
epochs. (a) Results for seed data generated with the conven-
tional method and (b) results for seed data generated with
the randomized method.
think that our deep learning based approach is useful
for increasing the accuracy of nonrigid registration.
We next evaluated the seed generator by com-
paring the conventional method with a randomized
method. We generated seed sets with random de-
formation vector fields, generated training sets, and
trained the GoogLeNet with 25 epochs to estimate
deformation directions for control points. Figure 5
shows the success rate r = s/t with different numbers
of epochs, where s is the number of correct estima-
tions and t is the total number of estimations. The
training accuracy and testing accuracy here are infer-
ence results for training sets and validation sets, re-
spectively. In this figure, the accuracy with our train-
ing sets reached success rate of 80%, but that with
random sets resulted in 70%. Thus, the proposed
method produced more realistic training sets as com-
pared with the randomized method. As for testing ac-
curacy, we found that ten epochs were sufficient to
saturate the accuracy of registration. Therefore, we
think that training phase can be completed in approx-
imately three hours on the latest GPU.
6 CONCLUSION
In this paper, we presented an automated method for
generating training set for image registration, aim-
ing at realizing nonrigid registration with deep learn-
ing. The proposed method generates enormous train-
ing sets from a small number of seed sets, i.e., de-
formation vector fields obtained with a conventional
method. This automated approach allows users to
minimize effort for collecting training sets required
for deep learning.
In experiments, we applied the proposed method
to registration of MR brain images. As seed sets,
we first obtained 30 deformation vector fields with
the conventional method. We then generated 169,890
training sets in 33 minutes from the initial seed sets
applied to 670,000 images. Given these training sets,
we trained the GoogLeNet and performed inference
with the trained network to estimate deformation di-
rections for every control point in the floating image.
As a result, the proposed method achieved precision
and recall of 80%, which were higher than 50% pro-
vided by the conventional method. Therefore, we
think that the proposed method is useful for realizing
image registration with deep learning.
Future work includes the development of the full
registration framework with supporting 3-D images; a
scheme that predicts both the deformation length and
direction is further required to realize full registration.
ACKNOWLEDGEMENTS
This study was supported in part by the Japan Soci-
ety for the Promotion of Science KAKENHI Grant
Numbers 15H01687, 16H02801 and 15K12008. We
are also grateful to the anonymous reviewers for their
valuable comments.
REFERENCES
Cheng, X., Zhang, L., and Zheng, Y. (2016). Deep simi-
larity learning for multimodal medical images. Com-
puter Methods in Biomechanics and Biomedical Engi-
neering: Imaging & Visualization, pages 1–5.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei,
L. (2009). Imagenet: A large-scale hierarchical image
database. In Proc. 22nd IEEE Conf. Computer Vision
and Pattern Recognition (CVPR’09), pages 248–255.
Ghosal, S. and Ray, N. (2017). Deep deformable registra-
tion: Enhancing accuracy by fully convolutional neu-
ral net. Pattern Recognition Letters, 94(15):81–86.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. MIT Press.
BIOIMAGING 2018 - 5th International Conference on Bioimaging
146

Hajnal, J. V., Hill, D. L., and Hawkes, D. J., editors (2001).
Medical Image Registration. CRC Press, Boca Raton,
FL.
Howard, A. G. (2013). Some improvements on deep con-
volutional neural network based image classification.
arXiv:1312.5402.
Ikeda, K., Ino, F., and Hagihara, K. (2014). Efficient accel-
eration of mutual information computation for non-
rigid registration using CUDA. IEEE J. Biomedical
and Health Informatics, 18(3):956–968.
Ino, F., Ooyama, K., and Hagihara, K. (2005a). A data
distributed parallel algorithm for nonrigid image reg-
istration. Parallel Computing, 31(1):19–43.
Ino, F., Tanaka, Y., Kitaoka, H., and Hagihara, K. (2005b).
Performance study of nonrigid registration algorithm
for investigating lung disease on clusters. In Proc. 6th
Int’l Conf. Parallel and Distributed Computing, Appli-
cations and Technology (PDCAT’05), pages 820–825.
Ioffe, S. and Szegedy, C. (2015). Batch normalization: Ac-
celerating deep network training by reducing internal
covariate shift. arXiv:1502.03167.
Klein, A., Andersson, J., Ardekani, B. A., Ashburner,
J., Avants, B., Chiang, M.-C., Christensen, G. E.,
Collins, D. L., Gee, J., Hellier, P., Song, J. H., Jenk-
inson, M., Lepage, C., Rueckert, D., Thompson, P.,
Vercauteren, T., Woods, R. P., Mann, J. J., and Parsey,
R. V. (2009). Evaluation of 14 nonlinear deformation
algorithms applied to human brain MRI registration.
NeuroImage, 46(3):786–802.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neu-
ral networks. In Proc. 26th Conf. Neural Information
Processing Systems (NIPS’12), pages 1097–1105.
Lee, S., Wolberg, G., and Shin, S. Y. (1997). Scattered data
interpolation with multilevel B-splines. IEEE Trans.
Visualization and Computer Graphics, 3(3):228–244.
NVIDIA Corporation (2017a). CUDA C Programming
Guide Version 8.0.
NVIDIA Corporation (2017b). cuDNN v5.1: GPU acceler-
ated deep learning.
Preferred Networks, inc. (2017). Chainer: A powerful, flex-
ible, and intuitive framework for neural networks.
Rohlfing, T. and Maurer, C. R. (2003). Nonrigid image reg-
istration in shared-memory multiprocessor environ-
ments with application to brains, breasts, and bees.
IEEE Trans. Information Technology in Biomedicine,
7(1):16–25.
Rueckert, D., Sonoda, L. I., Hayes, C., Hill, D. L. G., Leach,
M. O., and Hawkes, D. J. (1999). Nonrigid regis-
tration using free-form deformations: Application to
breast MR images. IEEE Trans. Medical Imaging,
18(8):712–721.
Shen, D. and Davatzikos, C. (2002). HAMMER: Hierar-
chical attribute matching mechanism for elastic regis-
tration. IEEE Trans. Medical Imaging, 21(11):1421–
1439.
Studholme, C., Hill, D. L. G., and Hawkes, D. J. (1999).
An overlap invariant entropy measure of 3D medical
image alignment. Pattern Recognition, 32(1):71–86.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Rabi-
novich, A. (2015). Going deeper with convolutions.
In Proc. 28th IEEE Conf. Computer Vision and Pat-
tern Recognition (CVPR’15), pages 1–9.
Vercauteren, T., Pennec, X., Perchant, A., and Ay-
ache, N. (2009). Diffeomorphic demons: Effi-
cient non-parametric image registration. NeuroImage,
45(1):S61–S72.
Wu, G., Kim, M., Wang, Q., Gao, Y., Liao, S., and
Shen, D. (2013). Unsupervised deep feature learning
for deformable registration of MR brain images. In
Proc. 16th Int’l Conf. Medical Image Computing and
Computer-Assisted Intervention (MICCAI’13), pages
649–656.
Wyman, B. T., Harvey, D. J., Crawford, K., Bernstein,
M. A., Carmichael, O., Cole, P. E., Crane, P. K., De-
Carli, C., Fox, N. C., Gunter, J. L., et al. (2013). Stan-
dardization of analysis sets for reporting results from
ADNI MRI data. Alzheimer’s & Dementia, 9(3):332–
337.
An Automated Method for Generating Training Sets for Deep Learning based Image Registration
147
