
Comparative Study of Depth-Image Matching with Steepest
Descendent and Simulated Annealing Algorithms
Hiroshi Noborio, Shogo Yoshida, Kaoru Watanabe, Daiki Yano and Masanao Koeda
Department of Computer Science, Osaka Electro-Communication University, Shijo-Nawate, Japan
Keywords: Simulated Annealing, Steepest Descendent, Organ-following Algorithm, Depth-Depth Matching.
Abstract: We have been developing a depth-depth matching-based organ-following algorithm for use in surgical
navigation. In this paper, we experimentally compare the steepest descendent and simulated annealing
algorithms under depth-depth matching. Our experiment can be performed in a real-life situation like practical
surgery as follows: the organ is artificially occluded by another object corresponding to a human body, and
surgery is performed on a real surgical bed beside two shadow-less lamps. In this research, in addition to the
algorithm comparison, we checked the effect of the placement of two infrared shielding filters, SL999 and
TS6080S. Based on the results, we could determine that the simulated annealing algorithm using the filter
TS6080S is the best.
1 INTRODUCTION
Many types of surgical navigation systems have been
developed in commercial and research fields. Nearly
all systems in the commercial field can be seen at the
"Medical expo" website (http://www.
medicalexpo.com/). Several surgical navigation
systems have been investigated in the research field
as well.
However, the intended targets of such systems are
the brain (Ferroli, 2013; Schulz, 2012) or the bone
(Blakeney 2011; Schnurr 2011). In almost all
navigation researches, the bone is completely fixed
by plastic components and consequently is not
deformed. In addition, the brain is almost entirely
confined by the skull and is not deformable except
when considering the well-known case of brain shift.
Therefore, the shape, position, and orientation of the
brain and its inner parts such as several kinds of
tumours, blood vessels, and nerves are precisely
calculated.
Therefore, conventional surgical navigations
systems do not consider large rotations/translations or
deformations in the targeted systems. For this reason,
we focus on the organ-following algorithm that
considers the deformation by using a liver with
arteries, veins, portal veins, and tumor, or brain with
a shift, and so on (Noborio, 2013; Noborio, 2016a;
Noborio 2016b). We compared several kinds of depth
cameras such as RICOH SV-M-S1, SICK V3310,
Microsoft Kinect v1 and v2, Intel RealSense, and
Euclid to get the deformation (the surface depth
image) of the liver and brain during a real-time
surgery. In this paper, we focused on following a real
organ and scalpel cavitron ultrasonic surgical
aspirator (CUSA) by its virtual organ and scalpel
CUSA after calibrating both coordinate systems. Our
previous papers addressed this agenda realistically by
attaching some cameras directly besides a surgical
bed in a real surgical room with two shadow-less
lamps covered by two types of infrared filters. In this
paper, while maintaining this realistic set-up, we
experimentally compare two types of search
algorithms—steepest descendent and simulation
annealing. In addition, during the comparison, we
tested the effect of placing two infrared shielding
filters SL999 and TS6080S around the two shadow-
less lamps. The results showed that the combination
of simulation annealing and the infrared filter
TS6080S is the best for following a real liver by its
virtual liver in a real surgical room with two shadow-
less lamps.
In section 2, we describe the real and virtual livers
used in our experiment, and then explain our key idea
of depth-image matching. In section 3, we first show
our surgical room for an obstructed liver, which is
equipped with two shadow-less lamps. Then, we
compare the steepest descendent and simulated
Noborio, H., Yoshida, S., Watanabe, K., Yano, D. and Koeda, M.
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms.
DOI: 10.5220/0006644200770087
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 1: BIODEVICES, pages 77-87
ISBN: 978-989-758-277-6
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
77
annealing algorithms. Finally, in section 4, we present
some concluding remarks.
2 DEPTH-IMAGE MATCHING
AND OVERLAPPING RATIO
BETWEEN REAL AND
VIRTUAL LIVERS
From 2014, we have been developing several kinds of
organ-following algorithms based on the steepest
descendent algorithm from a virtual world simulation
(Noborio, 2014a) to a real world experiment
(Noborio, 2015a). In addition, we tested these
algorithms in an experimental room and an actual
surgical room with zero, one, and two shadow-less
lamps. Moreover, we checked the usefulness of
placing infrared shielding filters, SL999 and
TS6080S (Noborio, 2017), on the lamp. Then, we
recently compared the classic steepest descendent
algorithm and the new simulated annealing algorithm
in an actual surgical room with two shadow-less
lamps enclosed by one of the two infrared filters,
SL999 and TS6080S.
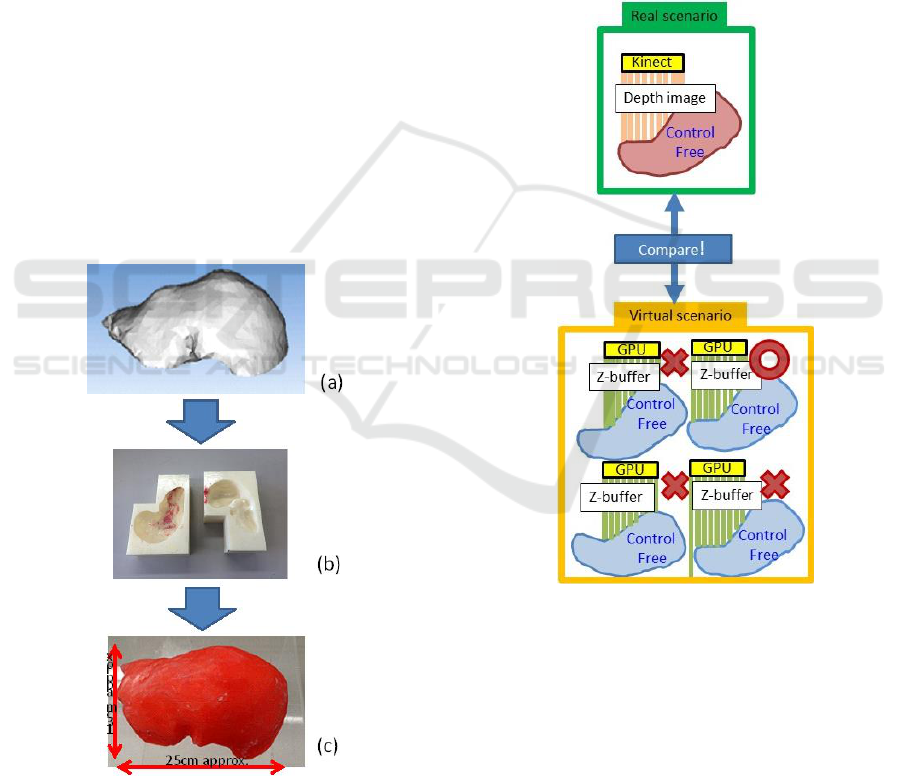
Figure 1: (a) Polyhedral liver in stereolithography STL
format, (b) its 3D-printed plastic template, and (c) plastic
liver pushed in the template.
In this chapter, we briefly explain our depth-depth
matching method used in two kinds of search
algorithms. The function value generated by the
depth-depth matching is used for the evaluation index
of the search algorithm. Then, we explain in detail the
steepest descendent and simulated annealing
algorithms. The steepest descendent algorithm does
not have a function for escaping from several local
minima. Therefore, it sometimes overlooks the
position/orientation where a virtual organ is
coincident with the real organ. To overcome this, we
prepared two kinds of randomized functions
(Noborio, 2016c). Contrary to this, the simulated
annealing algorithm includes the escaping function.
Therefore, we do not need to prepare such a function
additionally, which is an advantage.
Figure 2: Position/orientation of the virtual liver model is
analyzed to decrease the difference in its depth images and
that of the actual liver.
2.1 Fast Depth-Depth Matching
The most important function in our surgical
navigation is to track a virtual liver model (which is
displayed to users) against a real liver, and to match
the positions/orientations of the two liver samples at
any time (Figure 1).
In our research, a depth camera such as the Kinect
v2 captures the surface depth image of an organ.
Simultaneously, the graphics processing unit (GPU)
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
78

board on the PC generates the surface depth image of
the 3D virtual organ model. In Figure 2, we use the
depth-image matching between the real and virtual
worlds, whose positions and orientations were
completely calibrated in advance.
Figure 3: (a), (b) Search spaces containing 12 and 728
neighbouring candidates with six DOF space. (c),(d) Search
spaces containing 52 and 248 neighbouring candidates with
three translational DOF and three rotational DOF.
Each of the two kinds of proposed algorithms will
always seek a better positon/orientation of the virtual
liver against the positon/orientation of the real liver.
By shifting slightly each time within the six degrees-
of-freedom (DOF) space by (x, y, z, φ, θ, ψ), where
x, y, z are the coordinates in the Euclidean 3D space,
φ is the roll angle, θ is the pitch, and ψ is the yaw
(Figure 3), we can find a better neighbouring point
(positon/orientation) of the virtual liver whose
overlapping ratio is minimum, as explained later. As
mentioned in (Watanabe, 2015), searching the 6D
digitalized space is quite time consuming. For this
reason, we attempted to divide the 6D space into a 3D
translation space and a 3D rotation space whose
difference was changed from 1 to 3.
To obtain a function value (=overlapping ratio)
using our search algorithm, it is better to match the
real and virtual depth images for matching the real
and virtual 3D point clouds (Liu, 2006; Wu, 2015).
The reason is as follows:
(1) The number of pixel depths is quite less than the
number of points (Figure 4).
(2) The former needs the sum of non-combinational
distance differences (e.g., for 1024*1024 pixels)
but the latter requires the sum of combinatorial
shortest distance calculations (e.g., for
combination pairs between one million or more
points in some point cloud) (Liu, 2006; Wu,
2015).
Figure 4: Summed differences between the real and virtual
depth images based on pixels selected randomly in parallel
on the GPU.
2.2 Randomized Algorithm based on
Steepest Descendent
Using a selected search algorithm, we can match the
positions and orientations of the virtual liver model
and the real liver. As mentioned, we tested two kinds
of randomized algorithms in order to decrease the
differences between the depth images of the virtual
and real liver models.
First, we designed a randomized algorithm based
on steepest descendent (Figure 5), which is used in a
real surgical room with an infrared filter, (Noborio,
2017) as follows:
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms
79

Figure 5: Flowchart illustrating depth-depth matching in
our randomized algorithm, based on the steepest descendent
method.
Given an initial guess x
0
, the method always
computes a sequence of iterates x
k
among six-
dimensional digitalized space (three are the
translational degrees-of-freedom and the other
three are the rotational degrees-of-freedom).
At x
k
, the algorithm always selects a neighbor
whose randomized value f is minimized from
many digitalized neighbors whose distances are
1 and 2 as x
k+1
, as shown in Figure 3.
Figure 6: We randomly select a group of N pixels from each
image. Then, we calculate the minimum, median, or
average of the difference distribution between the real and
virtual depth images. Furthermore, we select the minimum,
median, or average of the evaluation values in M images.
Their randomizations escape from a local minimum in the
motion space in our steepest descendent.
In this algorithm, in order to escape from several
local minima in the search space, we use two kinds of
randomizations (Figure 6).
(1) For each image, M pixels are randomly selected,
and the minimum, median, or average of their
differences are calculated.
(2) For N images, we select their minimum, median,
or average calculated in (1) as the evaluation value
for the search.
(3) In (1) and (2), we set M and N to 10, 50, and 100,
respectively. This parameter selection is
important for each organ.
2.3 Simulated Annealing Algorithm
In this section, we explain our simulated annealing
algorithm as another type of randomized algorithm
(Watanabe, 2017). Simulated annealing (SA) was
first proposed in 1983 by Kirkpatrick et al.
(Kirkpatrick, 1983). It was fortunately rediscovered
in 1985 by Vlado Cerny (Cerny, 1985). SA is a
metaheuristic that finds global optimization in a large
search space. Steepest descent is a simple search
method. At each step, it selects the best neighbor of
the current point that is stuck at a local optimum and
often cannot find a global optimum. SA selects a
neighbor probabilistically and finds the global
optimum for a sufficiently long time.
A_search(ti)
t ← ti
p ← position/orientation of the virtual liver
model at time ti
f ←fitness (p)
f_best ← f
p_best ← p
while (t i+1 - t > 0)
randomly select a neighbor p’ of p such
that |p’i - pi | ≦ D for all i
f ' ← fitness(p’)
if f' < f_best then
f_best ← f '
p_best ← p’
if f' < f or
random(0,1) ≧ (t - ti) / Δt then
f_best ← f '
p_best ← p’
t ← t+1
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
80

Figure 7: A 2D virtual liver, whose pixel is depicted in blue,
is starting to overlap with its 2D real liver, whose pixel is
depicted in red, in the depth image. In our matching system,
the overlapping area in the XY plane is described by a set
of yellow pixels, and the overlapping area in the XYZ space
is described by a set of blue pixels.
The random function (0,1) returns a random value
from zero to one. The fitness (p’) in this algorithm
equals the randomized value f of the steepest
descendent and it indicates the difference between the
two depth images. The parameter D is half the width
of the area, which is a six-dimensional hypercube
with width 2D, in which a new position/orientation p’
is randomly generated. If D increases, the algorithm
can choose a more distant neighbor to avoid falling
into a local minimum.
In this algorithm, f' < f_best implies that we obtain
the global minimum and f' < f implies that we obtain
the local minimum. To escape from the local
minimum, we prepare random (0,1) ≧ (t−ti) / Δt.
Because Δt increases, the random value is selected to
avoid falling into a local minimum. The camera can
capture a part of the 3D liver through the circular
hole.
First, we overlap a real liver by its virtual one in
the 3D space by watching the pixel states generated
by the real and virtual depth differences (Noborio
2015b). In our matching system (Figure 7), the areas
projected from the real and virtual livers into the XY
plane along the Z-axis are shown by sets of red and
blue pixels, respectively. The pixel overlapped by real
and virtual livers in the XY plane is shown in yellow.
Furthermore, the pixel overlapped by the real and
virtual livers in the XYZ space is shown in blue.
Therefore, an operator first moves the virtual liver in
the horizontal XY space by eliminating the red and
blue pixels in order to generate all yellow pixels.
Then, we move the virtual liver in the vertical XYZ
space by eliminating the yellow pixels and
simultaneously generating blue pixels.
Figure 8: We compared two kinds of motion transcription
algorithms in a real surgical room with (a) one shadow-less
lamp (b) two shadow-less lamps covered by two kinds of
invisible light filters. In our navigation, a 3D liver is always
obstructed by a cardboard box with a circular hole.
3 COMPARATIVE
EXPERIMENTAL RESULTS
In this chapter, we experimentally compared the
steepest descendent and the simulated annealing
algorithms with depth-depth matching.
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms
81

Figure 9: The same rotational movement is applied for the
randomized steepest descendent (a) and simulated
annealing algorithms (b). Unfortunately, right and left are
opposite in all 2D RGB and depth images.
3.1 Experiments in a Real Surgical
Room
In this section, we present several realistic
experiments, which were conducted not in the
laboratory, but in a surgical operating room. The
experimental equipment is precisely described in
Figure 8. An acrylic plate of length 25 cm, width of
25 cm, and thickness of 2 cm was placed on the
operating table and a real liver was placed on the top
of the plate. Consequently, an operator could flexibly
move the liver. Movement of the liver was achieved
by moving the acrylic plate instead of moving the
actual liver itself.
Figure 10: The same translational movement is applied for
the randomized steepest descendent (a) and simulated
annealing algorithms (b). Unfortunately, right and left are
opposite in all 2D RGB and depth images.
Moreover, in order to conduct experiments
involving occlusion, a cardboard sheet containing a
hole of 10 cm in diameter was placed on the liver. The
surface of this corrugated board was painted in light
orange using a color spray to make it look like human
skin. The camera (Kinect v2) was attached to a
vertically movable robot. Therefore, it was able to
change its distance from the liver according to
different situations. The camera was placed
horizontally with respect to the operating table at a
height of 84 cm from the bottom of the actual liver.
The camera system was fixed by attaching metal
fittings to a metal rod. The metal rod was fixed to the
camera, which was attached to the robot. The distance
from the robot to the camera was set as 32 cm.
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
82

Figure 11: The light transmittance (a) and characteristics (b)
of infrared shielding filter SL999, and the light
transmittance (c) and characteristics (d) of infrared
shielding filter TS6080.
The initial positions of the real liver and the
virtual liver model were matched in the first
experimental procedure. Next, the real liver placed on
an acrylic board was covered with corrugated
cardboard. It was subjected to rotational and
translational movement within the corrugated
cardboard box. It was done to ensure that the range of
the liver visible from the hole of the cardboard did not
become less than half after the movement of the
model. In our usual experiments, we select the offset
value of 10 mm for real and virtual depth images
against the 3D real liver and its polyhedron with the
STL format in our surgical navigator. The model was
rotated 45° around the center of the z-axis during the
rotational movement (Figure 9), and it was moved by
5 cm in the y-axis direction during the parallel
movement (Figure 10). Each co-ordinate axis is
selected as shown in Figure 8.
Figure 12: (a) Virtual and real RGB images, difference
between the real and virtual depth images, and the graph of
the overlapping ratio in the randomized steepest descendent
algorithm, and (b) real RGB image and real and virtual
depth images and their difference in the simulated
annealing algorithm.
3.2 Comparison of Rotational
Following Quality of the Two
Algorithms
In this section, we compare the depth-depth
matching-based organ-following algorithms for
several surgical rotation operations. We use the
steepest descendent (randomized) and simulated
annealing algorithms as the organ-following scheme.
Each algorithm always overlaps a virtual liver with its
real liver. The quality of overlap is evaluated by the
overlapping ratio explained in the previous section. It
is individually checked for two lamps without any
infrared shielding filter, with the SL999 or with the
TS6080S (Figure 11).
The purpose behind using SL999 or TS6080S is
that the depth camera itself uses infrared illumination.
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms
83

Figure 13: (a)(b) Comparative results of rotational liver operation between the randomized steepest descendent and simulated
annealing algorithms under two shadow-less lamps without any infrared shielding filter. (c)(d) Comparative results of
rotational liver operation between the randomized steepest descendent and simulated annealing algorithms under two shadow-
less lamps with the infrared shielding filter SL999. (e)(f) Comparative results of rotational liver operation between the
randomized steepest descendent and simulated annealing algorithms under two shadow-less lamps with the infrared shielding
filter TS6080S.
Kinect v2 gets the depth images by the Time of
Flight (TOF) method of projecting and receiving all
infrared rays. Therefore, the camera strictly tries to
gather the infrared rays of wavelength between 700
nm and 1000 nm. However, our shadow-less lamp
projects the infrared rays of wavelength under 400 nm
and over 1000 nm. Using our infrared filter SL999 or
TS6080S, the ultra-violet light whose wavelength is
between 10 nm and 400 nm is completely eliminated,
but the visible light whose wavelength is between 400
nm and 700 nm is not eliminated. In addition to this,
the infrared light and the radio waves over 1000 nm
are partially passed.
In our navigation system, we prepared a different
set of windows for the two algorithms, as described
in Figure 12. The reason is that the steepest
descendent (randomized) algorithm was formulated
during 2013–2015. In contrast, the simulated
annealing algorithm was independently developed
during 2016–2017.
However, in our navigation system, experimental
data such as sequences of real RGB images and depth
images during several kinds of surgical operations in
a real surgical room were completely memorized in
the PC under no infrared shielding filter. The reason
is as follows: a surgical room is always used by many
doctors, students, and researchers for surgeries,
lectures, and researches. Therefore, our experimental
data such as the sequences of lighting conditions
during surgeries, which were obtained from the surgi-
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
84

Figure 14: (a)(b) Comparative results of translational liver operation between the randomized steepest descendent and
simulated annealing algorithms under two shadow-less lamps without any infrared shielding filter. (c)(d) Comparative results
of translational liver operation between the randomized steepest descendent and simulated annealing algorithms under two
shadow-less lamps with the infrared shielding filter SL999. (e)(f) Comparative results of translational liver operation between
the randomized steepest descendent and simulated annealing algorithms under two shadow-less lamps with the infrared
shielding filter TS6080S.
cal room, are quite valuable. For this reason, all the
experimental information pertaining to the surgical
room was completely memorized and the algorithm
performance by varying several kinds of parameters
was checked again against the experimental
information in our laboratory. Therefore, we
compared two or more types of algorithms for the
same sequences of lighting.
A real liver is moved rotationally, and then its
virtual liver model follows the real liver. The same
rotational movement is used for the randomized
steepest descendent and simulated annealing
algorithms (Figure 9). As shown in Figure 13, the
initial overlapping ratio is over 97% (nearly equals
100%). When moved rotationally, as illustrated in
Figure 9, a variation in the overlapping ratio is
observed during the operation shown in Figure 13. In
almost all cases, the overlapping ratio obtained in the
simulated annealing algorithm was always better than
that achieved in the randomized steepest descendent
algorithm. Moreover, the overlapping ratio generated
by two lamps with TS6080S was always better than
with the other filter. As a result, the simulated
annealing algorithm using the filter TS6080S is
experimentally found to be the best with regard to the
rotational operation in liver surgery.
3.3 Comparison of Translational
Following Quality of the Two
Algorithms
A real liver is moved translationally, and then its vir-
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms
85

tual liver model follows the real liver. The same
translational movement is applied to the randomized
steepest descendent and simulated annealing
algorithms (Figure 10).
As shown in Figure 14, each overlapping ratio is
over 97% (nearly equals 100%). The variation in the
overlapping ratio under translational movement
(Figure 10) is shown in Figure 14. In almost all cases,
the poorest overlapping ratio obtained in the
simulated annealing algorithm is clearly better than
that obtained in the randomized steepest descendent
algorithm. Contrary to this, the overlapping ratios
generated by two lamps without any filter are almost
the same in the simulated annealing algorithm. As a
result, with respect to the translational operation of
liver surgery, the simulated annealing algorithm is
better but using the filters is experimentally
meaningless.
4 CONCLUSIONS
In this paper, we have given several comparative
results of depth-depth-image matching-based organ-
following algorithms. Compared with our previous
laboratory experiments, or without the addition of a
shadow-less lamp, this experiment can be applied to
real-life situations such as practical surgery as
follows: the organ is artificially occluded by another
object, and the surgical operation is performed on a
real surgical bed beside two shadow-less lamps. In
this research, in addition to the comparison of two
kinds of algorithms, we checked the usefulness of two
infrared shielding filters, SL999 and TS6080S. Based
on the results, we could determine that the simulated
annealing algorithm using the filter TS6080S is the
best.
At present, we are looking forward to using an
infrared filter, which can pass light of wavelength
between 700 nm and 1000 nm. In the near future, we
will try to verify the rotational or translational
following stability of our algorithm by using more
effective infrared shielding filters.
ACKNOWLEDGEMENTS
This study was partly supported by the 2014 Grants-
in-Aid for Scientific Research (No. 26289069) from
the Ministry of Education, Culture, Sports, Science
and Technology, Japan. The study was also supported
by the 2014 Cooperation Research Fund from the
Graduate School at Osaka Electro-Communication
University.
REFERENCES
Ferroli, P., Tringali, G., Acerbi, F., Schiariti, M., Broggi,
M., Aquino, D., Broggi, G., 2013. Advanced 3-
dimensional planning in neurosurgery, Neurosurg
Suppl. 1, pp.A54–A62. doi:10.1227/NEU.0b013e3182
748ee8.
Schulz, C., Waldeck, S., Mauer, M., 2012. Intraoperative
image guidance in neurosurgery: development, current
indications, and future trends, Radiol Res Pract
197364. doi:10.1155/2012/197364.
Blakeney, G., Khan, J., Wall, J., 2011. Computer-assisted
techniques versus conventional guides for component
alignment in total knee arthroplasty: a randomized
controlled trial, J Bone Joint Surg Am. 93, pp.1377–
1384. doi: 10.2106/JBJS.I.01321.
Schnurr, C., Eysel, P., König, P., 2011. Displays mounted
on cutting blocks reduce the learning curve in navigated
total knee arthroplasty, Comput Aided Surg. 16:249–
256. doi: 10.3109/10929088.2011.603750.
Noborio, H., et al., 2013. A Fast Surgical Algorithm
Operating Polyhedrons Using Z-Buffer in GPU. In 9th
Asian Conference on Computer Aided Surgery, Waseda
University, Tokyo Japan, pp.110-111.
Noborio, H., et al., 2016a. Fast Surgical Algorithm for
Cutting with Liver Standard Triangulation Language
Format Using Z-Buffers in Graphics Processing Unit.
Computer Aided Surgery, DOI:10.1007/978-4-431-
55810-1, pp.127-140.
Noborio, H., et al., 2016b. Depth Image Matching
Algorithm for Deforming and Cutting a Virtual Liver
via its Real Liver Captured by Kinect v2. In IWBBIO
2016, LNBI 9656, pp.196-205, DOI: 10.1007/978-3-
319-31744-1_18.
Watanabe, K., et al., 2015. A New 2D Depth-Depth
Matching Algorithm whose Translation and Rotation
Freedoms are Separated. In ICIIBMS2015, Track 3:
Bioinformatics, Medical Imaging and Neuroscience,
OIST, Okinawa Japan, pp.271-278, 2015.
Liu, Y, 2006. Automatic registration of overlapping 3D
point clouds using closest points. Journal of Image and
Vision Computing, Vol. 24, No. 7, pp.762-78.
Wu, F., Wang, W., Lu, Q., Wei, D., Chen, C., 2015. A new
method for registration of 3D point sets with low
overlapping ratios. In 13th CIRP Conference on
Computer Aided Tolerancing, pp.202-206.
Noborio, H., et al, 2016c. Tracking a Real Liver using a
Virtual Liver and an Experimental Evaluation with
Kinect v2. In IWBBIO 2016, LNBI 9656, pp.149-162,
DOI: 10.1007/978-3-319-31744-1_14.
Noborio, H., et al. 2014. Motion Transcription Algorithm
By Matching Corresponding Depth Image and Z-
buffer, In 10th Anniversary Asian Conference on
Computer Aided Surgery, pp.60-61.
BIODEVICES 2018 - 11th International Conference on Biomedical Electronics and Devices
86

Noborio, H., et al. 2015a. Experimental Results of 2D
Depth-Depth Matching Algorithm Based on Depth
Camera Kinect v1. Journal of Bioinformatics and
Neuroscience, Vol.1, No.1, pp.38-44.
Noborio, H., et al., 2017. Algorithm Experimental
Evaluation for an Occluded Liver with/without
Shadow-Less Lamps and Invisible Light Filter in a
Surgical Room. In HCI 19, Lecture Notes in Computer
Science book series (LNCS, volume 10289), pp.524-
539, DOI: 10.1007/978-3-319-58637-3_41.
Watanabe, K., et al, 2017. A New Organ-Following
Algorithm Based on Depth-Depth Matching and
Simulated Annealing, and Its Experimental Evaluation.
In HCI 19, Lecture Notes in Computer Science book
series (LNCS, volume 10289), DOI:10.1007/978-3-
319-58637-3_47 pp.594-607.
Kirkpatrick, S., Gelatt, D., Vecchi, P., 1983. Optimization
by simulated annealing. Science 220 (4598), pp.671-
680. DOI:10.1126/science.220.4598.671. JSTOR
1690046. PMID 17813860.
Cerny, V., 1985. Thermodynamical approach to the
traveling salesman problem: An efficient simulation
algorithm. Journal of Optimization Theory and
Applications, Vol.45, No.1, pp.41-51.
DOI:10.1007/BF00940812.
Noborio, H, et al. 2015b. Image-based Initial
Position/Orientation Adjustment System between Real
and Virtual Livers," Jurnal Teknologi, Medical
Engineering, Vol. 77, No.6, pp. 41-45,
DOI:10.11113/jt.v77.6225, Penerbit UTM Press, E-
ISSN 2180-3722.
Comparative Study of Depth-Image Matching with Steepest Descendent and Simulated Annealing Algorithms
87
