
geneEX
A Novel Tool to Assess Differential Expression
from Gene and Exon Sequencing Data
Orazio M Scicolone
∗
, Giulia Paciello
∗
and Elisa Ficarra
Department of Control and Computer Engineering, Politecnico di Torino, Torino, Italy
Keywords:
RNA-Sequencing, Differentially Expressed Genes, Exon Analysis, Table of Counts.
Abstract:
The widespread of Next Generation Sequencing technologies accounted in recent years for the possibility to
evaluate gene expression with great accuracy. Moreover, it allowed assessing differential gene expression
among biological conditions with high sensitivity. However, state-of-the-art bioinformatics methodologies for
differential gene expression evaluation from RNA Sequencing data still suffer from several drawbacks such
as reduced specificity. In this paper we propose geneEx, a novel methodology and tool for differential gene
expression evaluation from RNA Sequencing reads. By combining gene and exon expression evaluation and
BioMart information, geneEX provides users with annotated lists of highly reliable differentially expressed
genes. The results obtained in Sequencing Quality Control dataset proven the importance of a novel approach
to lower False Positive predictions from current methodologies and the strength of the proposed methodolog-
ical approach to increase the sensitivity of differentially expressed gene identification.
1 INTRODUCTION
The advent of Next Generation Sequencing (NGS)
technologies dramatically reshaped genomics and
cancer genomics, allowing to produce huge amounts
of sequencing data with reduced per-base costs. Older
platforms such as microarrays were largely replaced
in the last decades by NGS techniques (Lee et al.,
2013). These techniques were exploited in several
contexts and with different purposes, such as the char-
acterisation of novel genomes, the deepening of par-
tially known genomic structures, or the identification
of new variants at the base pair resolution.
Moreover, massive cDNA sequencing, also known
as RNA-Sequencing (RNA-Seq), was adopted to
carefully analyse and quantify transcriptomes, al-
lowing to discriminate differentially expressed genes
(DEGs) among biological conditions. To this aim,
several algorithms working on NGS data were de-
veloped. These algorithms implement different ap-
proaches for data normalization and DEG identifica-
tion, as widely discussed in (Young et al., 2012). Six
of these methods, i.e. Cuffdiff (Trapnell et al., 2013),
edgeR (Robinson et al., 2010), DESeq (Love et al.,
2014), PoissonSeq (Li et al., 2012), baySeq (Hardcas-
tle and Kelly, 2010) and limma (Smyth, 2005), were
∗
co-first-author
recently compared in SEQC (DeLuca et al., 2012) and
ENCODE (Consortium et al., 2004) datasets consid-
ering, as examples, their normalisation and specificity
performance. Overall, no specific method was proven
to be the best solution in all the comparisons (Rapa-
port et al., 2013).
To the light of this consideration, as stated in (Ra-
jkumar et al., 2015), the combined adoption of dif-
ferent algorithms for differential gene expression as-
sessment is highly advisable to achieve good sensi-
tivity. However, it has to be considered that higher
the number of tools adopted higher both the compu-
tational costs and the amount of False Positive (FP)
predictions obtained in output.
To overcome limitations proper of state-of-the-art
methodologies for DEG identification, we propose
a novel methodology and tool named geneEX. ge-
neEX comes in the form of an R package that can
be easily integrated within all of the bioinformatics
pipelines working on RNA-Seq data. It performs
DEG assessment by processing and integrating the
results from three widely adopted R packages, i.e.,
DESeq2, edgeR, and DEXSeq. Specifically, the first
two methods are exploited by geneEX to perform
expression evaluation at the gene level, whereas the
last one to make exon expression assessment. ge-
neEX implements a series of elaboration and filtering
174
Scicolone, O., Paciello, G. and Ficarra, E.
geneEx - A Novel Tool to Assess Differential Expression from Gene and Exon Sequencing Data.
DOI: 10.5220/0006657501740181
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 3: BIOINFORMATICS, pages 174-181
ISBN: 978-989-758-280-6
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

stages which allow to shrink down the list of DEGs
from current methodologies, focusing on the more re-
liable ones. By integrating annotations from BioMart
database (Durinck et al., 2005), geneEX provides bi-
ologists with a series of information useful to fur-
ther prioritise true DEGs. Moreover, its analysis can
be easily triggered according to both user needs and
computational resources. The novel geneEX will be
released as soon as possible as a Bioconductor pack-
age (Huber et al., 2015).
We assessed the need for a novel DEG analy-
sis tool and geneEX performance in the Sequencing
Quality Control (SEQC) dataset (Shi et al., ; Shi et al.,
2006). Results from these analyses proven the impor-
tance of the filtering stages implemented within ge-
neEX to lower FPs, thus increasing the specificity of
the detection and providing users with highly reliable
DEG candidates.
2 MATERIALS AND METHODS
2.1 Dataset
Data exploited in our study was downloaded from
SRA database with accession codes SRX333347-
SRX333356. The considered dataset is part of SEQC
study and comprises two groups of samples, which
will be referred to as Group 1 and Group 2 in the fol-
lowing. Specifically, the five samples belonging to
the first group are replicates of the Stratagene Uni-
versal Human Reference RNA (UHRR) that contains
the RNA from ten human cell lines with 2% by vol-
ume of synthetic RNAs from the External RNA Con-
trol Consortium (ERCC) mix1. Conversely, Group
2 comprises five replicates of the Ambion’s Human
Brain Reference RNA (HBRR) with 2% by volume
of ERCC mix2. ERCC spike-in control mixes are sets
of 92 250-2000 nt (nucleotide) long polyadenylated
transcripts from the ERCC plasmid reference library.
These spike-in sequences can be further grouped into
four subgroups, with well defined molar concentra-
tions in mix1 and mix2.
2.2 Read Alignment and ene/xon Count
Calculation
According to (Rapaport et al., 2013) study, paired-end
reads were aligned using tophat2 (Kim et al., 2013)
on a reference sequence comprising both human
chromosomes and ERCC spike-in nucleotide series.
UCSC gene and ERCC spike-in counts were retrieved
from tophat2 output files by using HTSeq (Anders
et al., 2015). Tophat2 and HTSeq parameters were
tuned as stated in (Rapaport et al., 2013). Exon counts
were obtained by running DEXSeq dexseq count.py
program with default parameters. The gtf annotation
file exploited during tophat2, HTSeq and DEXseq
analyses comprises both spike-in and UCSC gene se-
quences, according to (Rapaport et al., 2013).
2.3 geneEX Workflow
geneEX tool is an automatic, parallel and highly cus-
tomizable R module for differential gene expression
analysis that works on RNA-Seq data.
It is built on-top of three widely adopted state-of-
the-art Bioconductor (Gentleman et al., 2004) pack-
ages for differential gene expression evaluation, i.e.,
DESeq2 (Love et al., 2014), edgeR (Robinson et al.,
2010) and DEXSeq (Li et al., 2015). These packages
provide a method to test for differential gene/exon ex-
pression among biological conditions by exploiting a
statistical model to describe data distribution. Specif-
ically, DESeq2 and DEXSeq model data using a neg-
ative binomial distribution, whereas edgeR fits data
using the Poisson distribution.
The input data of these algorithms is represented
by the so called tables of counts. The table of
counts reports, for each gene or exon in the genome,
the number of reads (i.e., the elementary sequences
output of Next Generation Sequencing machines)
mapped on it. In more detail, edgeR was designed
to identify differentially expressed genes or exons,
DESeq2 to retrieve differentially expressed genes and
DEXSeq to identify exon usage biases among condi-
tions.
geneEX elaborates and combines the results from
these three tools to provide users with a reduced list
of highly reliable DEGs. Moreover, geneEX anno-
tates these genes with a series of information from
BioMart (Smedley et al., 2015) database, facilitating
they further prioritization by biologist and clinician
investigation.
Specifically, geneEX makes use of DESeq2 and
edgeR to test for differential genes, whereas of
DEXSeq to further prioritized these candidates based
on exon expression data.
geneEX workflow is depicted in Figure 1 and
detailed in the following. Rectangular white boxes
identify those activities implemented by state-of-the-
art bioinformatics algorithms, whereas the grey ones
those performed by ad-hoc designed R scripts. Con-
versely, yellow, pink and orange irregular shapes re-
port respectively on the input data, the output files
from the differential analysis and the results from
Rlog normalisation.
geneEx - A Novel Tool to Assess Differential Expression from Gene and Exon Sequencing Data
175

Figure 1: geneEX flowchart.
Starting point of geneEX analysis is the setting of
the tool parameters within a configuration file (block
A in Figure 1).
Users have to specify i) the path of the folder
that stores the input table of counts organized ac-
cording to the biological condition, ii) the path of
an annotation file that is exploited by DEXSeq to
create a DEXSeq object, iii) the number of threads
to be launched during the analysis, iv) the statisti-
cal measures, i.e., p-value (pval), adjusted p-value
(adj-pval), False Discovery Rate (FDR) or logarith-
mic Fold Change (log
2
FC) and their values to be
adopted by DESeq2, edgeR and DEXSeq to filter out
data, and iv) the number of differentially expressed
exons from DEXSeq results to be considered to fur-
ther filter out data. As shown in block C of Figure
1, the input gene/exon table of counts are processed
by geneEX to generate DESeq2, edgeR and DEXSeq
objects. These objects represent the input required
by DESeq2, edgeR and DEXSeq to perform differen-
tial gene/exon expression analysis. Additionally, gene
counts are normalized by DESeq2 RLog Transform
function (block B in Figure 1) which converts the raw
counts into log2-scale data. This to minimise the dif-
ferences among samples for low expressed genes and
to normalise data based on library sizes. RLog trans-
formation represents a fundamental preliminary step
for different analysis techniques such as clustering or
PCA (Love et al., 2014). These methods can be eas-
ily integrated within the novel implemented package,
making geneEX a very versatile tool.
The next phase of geneEX, shown in block D of
Figure 1, consists in the execution of DESeq2, edgeR
and DEXSeq algorithms. The output of both DESeq2
and edgeR runs is a list of genes, where each gene
comes with several statistical scores such as pval,
adj-pval, FDR and log
2
FC, that describe the confi-
dence in assessing differential gene expression. Sim-
ilarly, these results are provided in terms of exons by
DEXSeq. These three output files (i.e., DESeq2 re-
sults, DEXSeq results and edgeR results in Figure 1)
are saved in CSV files.
Step E implements the first filtering activity to be
performed on the output lists from DESeq2, edgeR
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
176

and DEXSeq tools. Specifically, results are filtered
according to the fixed thresholds and saved into CSV
files. In F, the gene lists from DESeq2 and edgeR
analyses are further elaborated, by retaining those
genes with at least a given number of exons (fixed in
the parameter setting activity) that are with high prob-
ability differentially expressed (according to DEXSeq
analysis and the adopted filtering threshold). Even in
this case results are provided to users in CSV format.
It is worth noting that users are let free to select the
statistical measure (and its value) to filter data based
on exon expression. This allows to implement a fil-
tering step that considers the specific protocol adopted
for RNA extraction (in most of cases poly(A) selection
or rRNA depletion). Finally, in step G, those genes
shared by the last two files are extracted and saved in
a CSV file. The whole lists of genes contained in both
the intermediate and final output files are annotated
in the different processing steps by querying Biomart
database(Smedley et al., 2015). Thus, each candidate
gene will be provided to the user with a series of infor-
mation such as the chromosome to which it belongs
and the relative chromosome band, its start and end
positions on the chromosome and the HUGO Gene
Nomenclature Committee (HGNC) symbol. All this
information is essential to further prioritise true dif-
ferentially expressed genes based on the expertise of
biologists and clinicians.
3 RESULTS AND DISCUSSION
We used ERCC spike-in synthetic sequences to assess
the performance of DESeq2 and edgeR tools, on-top
of which geneEX is built. These results allowed to
point out the need for ad-hoc filtering strategies to fo-
cus on a reduced list of highly reliable DEGs to be fur-
ther deepened by wet-lab experiments. Conversely,
non-synthetic reads from SEQC (UHRR and HBRR
datasets) were exploited to test the whole geneEX al-
gorithm and to discuss the results it provided.
3.1 ERCC Spike-in Analysis
ERCC spike-in synthetic sequences cannot be di-
vided into exons. Thus, reads deriving from their se-
quencing are not suitable for geneEX analysis, which
comprises DEXSeq processing. However, ERCC
dataset can be used as a benchmark to evaluate both
edgeR and DESeq2 performance. We ran DESeq2
and edgeR with default parameters comparing Group
1 and Group 2. Specifically, we focused on those
spike-in sequences not differentially expressed since
characterised by equal molar concentration in both
mix1 and mix2 (ERCC subgroup ID: B). The adj-
pvals provided by DESeq2 and the FDRs from edgeR
for these 23 not differentially expressed ERCC se-
quences were used to build the respective Receiver
Operating Characteristic (ROC) curves and to calcu-
late the relative Area Under Curves (AUCs). The
blue and yellow ROC curves of Figure 3 report on
DESeq2 and edgeR results respectively. Both tools
ensured sensitivity levels close to 0.8 with a speci-
ficity of about 0.56 when thresholds equal to 0.034
and 0.01 are applied to the adj-pvals from DESeq2
and the FDRs from edgeR. These results confirmed
the importance of ad-hoc filtering strategies based
on statistical measures such as the pval, to obtain
satisfactory sensitivity and specificity levels. More-
over, we assessed DESeq2 and edgeR capability to
correctly identify ERCC differentially expressed se-
quences (ERCC subgroup IDs: A, C and D) when ap-
plying different adj-pval and FDR thresholds. These
sequences were selected according to a log
2
FC6= 0
between Group 1 and Group 2. 69 out of 92 ERCC
sequences were identified as differentially expressed.
Two analyses were performed, using as input for DE-
Seq2 and edgeR run a table of counts containing i)
ERCC sequences only or ii) the whole list of hg19
annotated genes and the complete set of ERCC se-
quences. This twofold analysis allowed us to discuss
both the obtained results in terms of False Positive
(FP), False Negative (FN), True Negative (TN) and
True Positive (TP) predictions and the strength of the
statistical model applied by DESeq2 and edgeR. Fig-
ure 2 reports on these results. Specifically, each Venn
diagram depicts in the circle labelled as ERCC the
number of differentially expressed sequences defined
in ERCC experiment, whereas in the circles named
DESeq2 and edgeR the amount of differentially ex-
pressed sequences identified by DESeq2 and edgeR
from RNA-Seq data. The different subfigures in the
first row of Figure 2 are relative to the former analy-
sis, whereas the second row reports on the latter anal-
ysis.
With reference to the first analysis (that involved
ERCC sequences only), this allowed to carefully eval-
uate the performance of DESeq2 and edgeR in terms
of sensitivity and specificity. With the sensitivity cal-
culated as the ratio between the amount of TPs and the
sum of TP and FN predictions, whereas the specificity
as the ratio between TNs and the sum of TN and FP
predictions. This analysis allowed also to assess the
convenience of a DESeq2-edgeR combined approach
for DEG identification. Specifically, sequences iden-
tified by one or both tools and shared with ERCC set
are TPs, whereas those not shared with ERCC set are
FP predictions. Sequences included in ERCC dataset
geneEx - A Novel Tool to Assess Differential Expression from Gene and Exon Sequencing Data
177

Figure 2: Consensus among in-silico and wet-lab methodologies on ERCC spike-in differentially expressed sequence identi-
fication. Subfigures A, B and C report on the results obtained when considering ERCC sequences only, whereas Subfigures
D, E and F depict those provided by the adopted methodologies on the whole list of hg19 annotated genes and the complete
set of ERCC sequences.
only are FN predictions, and those not contained in
ERCC set (non differentially expressed) and not iden-
tified by the tools are TNs. These data are shown in
Table 1.
The different rows of Table 1 report on DESeq2,
edgeR and DESeq2-edgeR results respectively. The
adoption of DESeq2 ensured the best results in terms
of sensitivity and specificity. However, it has to be
considered that the number of sequences involved in
this analysis is very reduced, far from the number of
genes considered in standard experiments. As conse-
quence, the reduced number of FP predictions is not
meaningful, making useless an approach that com-
bines DESeq2 and edgeR tools since negatively im-
pacting on the sensitivity while not improving speci-
ficity. However, as clearly stated in (Rapaport et al.,
2013), in real cases the number of FPs is gener-
ally very huge, calling for ad-hoc filtering procedures
to shrink down their numbers. This aspect clearly
emerged in the second analysis, whose results are re-
ported in the second row of Figure 2. The lack of
a complete wet-lab validation for all genes involved
in the experiment makes impossible the calculation
of the overall number of FP, TP, FN and TN predic-
tions. However, the number of TP ERCC sequences
identified by DESeq2 and edgeR is higher than that
reported as result of the first analysis. Confirming the
importance of a statistically meaningful input to build
the probability distribution exploited by DESeq2 and
edgeR tools. Moreover, with high likelihood, not all
the genes labelled as differential by DESeq2, edgeR
or DESeq2-edgeR are in reality TPs.
This confirms the need for an accurate selection of
the statistical measure and its value to be adopted to
lower FP rates while maintaining satisfactory sensi-
tivity levels, keeping in mind that too strict thresholds
can negatively impact on sensitivity while poorly im-
proving specificity. By exploiting exon analysis, ge-
neEX accounts for the possibility to adopt more re-
laxed thresholds that preserve sensitivity while lower-
ing the number of FPs.
3.2 SEQC Non Synthetic Data Analysis
In (Canales et al., 2006), three different quantitative
gene expression measurement technologies were as-
sessed, i.e. TaqMan Gene Expression Assay, Stan-
dardized RT (Sta)RT-PCR assay and QuantiGene as-
say, and the obtained results compared with those
from DNA microarray platforms. Specifically, Taq-
Man assays were performed on 997 genes. We ex-
ploited this set of genes to evaluate geneEX per-
formance, defining as DEGs those genes having a
log
2
FC > 0.5 or < −0.5 between Group 1 and Group
2 according to qRT-PCR analysis. In more detail, our
analysis comprised four replicates from the UHRR
for Group 1, whereas four replicates of the HBRR
for Group 2. 30 out 997 genes were excluded from
our experimental setup because not found in BioMart
database. Based on the adopted log
2
FC, 742 genes
were marked as differentially expressed, whereas 225
as a not differential.
We ran DESeq2, edgeR and geneEX on the 967
genes from TaqMan assays imposing default param-
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
178
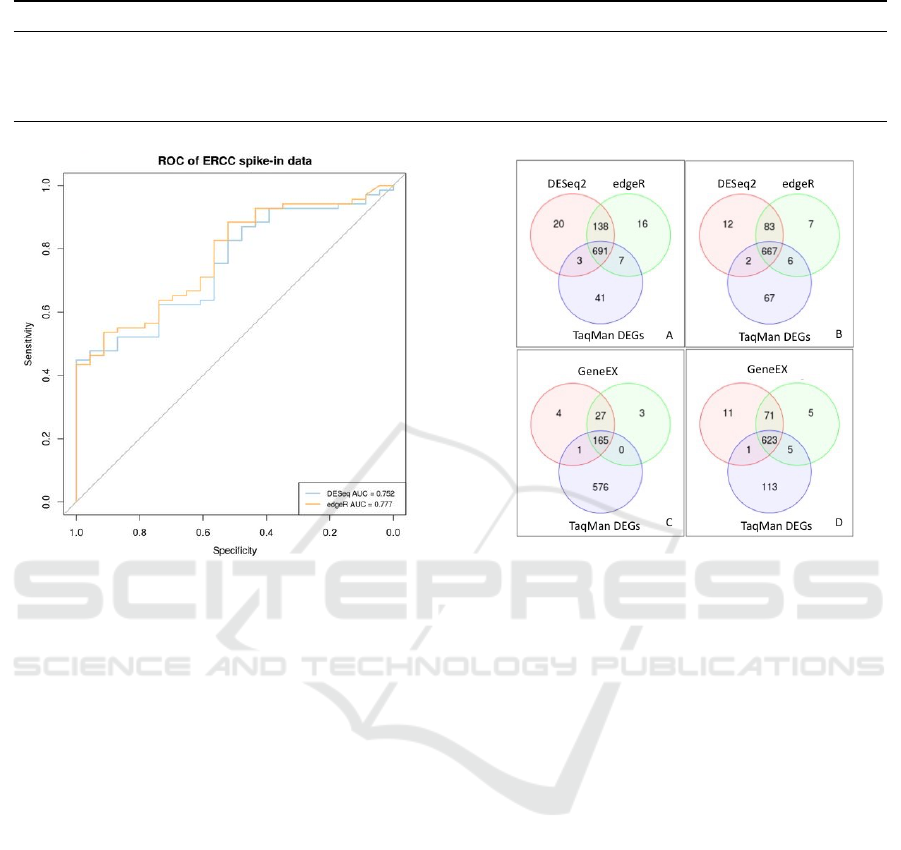
Table 1: Sensitivity/Specificity levels of DESeq2, edgeR and DESeq2-edgeR .
FDR
edgeR
/adj-pval
DESeq2
=0.1 FDR
edgeR
/adj-pval
DESeq2
=0.05 FDR
edgeR
/adj-pval
DESeq2
=0.01
Sensitivity Specificity Sensitivity Specificity T Sensitivity Specificity
DESeq2 73.9% 47.8% 72.4% 60.9% 63.8% 73.9%
edgeR 73.9% 34.8% 66.7% 34.8% 53.6% 47.8%
edgeR ∩ DESeq2 66.6% 47.8% 63.8% 60.9% 53.6% 73.9%
Figure 3: ROC curves of DESeq2 and edgeR tools.
eters. The obtained results were analysed to assess
both the specificity/sensitivity of these methods when
used singly and those of a DESeq2-edgeR combined
approach. Specifically, two different statistical mea-
sures were adopted to identify the DEGs, i.e. the pval
and the log
2
FC. In the first analysis, based on pval
scores, we used as filtering threshold for DESeq2,
edgeR and geneEX, a value of 0.05. With reference
to geneEX, this threshold was applied to the all the
filtering stages implemented by the tools on-top of
which it is built (i.e., Block E in Figure 1). Simi-
larly, in the second analysis, a log
2
FC of 0.5 was im-
posed to focus on highly reliable DEGs. Figure 4 re-
ports on the number of DEGs shared among the com-
pared in-silico methodologies and the TaqMan assay
experimental results. Specifically, Subfigures 4.A and
4.C show respectively the amount of DEGs identi-
fied by TaqMan assay, DESeq2 and edgeR (Subfig-
ure A) or by TaqMan assay and geneEX (Subfigure
C), when imposing the pval filtering threshold. Sim-
ilarly, Subfigures 4.B and 4.D report on the num-
ber of genes identified by the same methodologies
when applying the log
2
FC threshold. With reference
to Subfigures 4.A and 4.B, the adoption of a differ-
ent statistical measure for data filtering, strongly im-
pacted on the obtained results. Indeed, by filtering us-
ing a pval threshold we obtained a higher percentage
Figure 4: Consensus among in-silico and wet-lab method-
ologies on UHRR and HBRR DEGs identification. Subfig-
ures A and C report on the amount of DEGs provided by the
adopted approaches when using the pval as statistical mea-
sure to filter data. Similarly, Subfigures B and D depict the
results provided by the same methodologies when filtering
according to a log
2
FC threshold.
of TP predictions, but at the same time a significant
amount of FP ones. Moreover, it has to be noticed
that, in both cases, even DESeq2-edgeR combined ap-
proach resulted in conspicuous amounts of FPs. ge-
neEX tries to lower the number of FPs by evaluating
exon expression data from DEXSeq analysis. As ex-
pected, the introduction of an additional filtering step,
negatively impacted on the number of identified true
DEGs. Indeed, when considering geneEX pval and
log
2
FC thresholds, we lost respectively about 54%
and 4.5% of TPs with respect to DESeq2-edgeR com-
bined approach results. However, geneEX analysis
also lowered the number of FPs, as desired. Account-
ing for a decrease of about 11.5% and 1.2% when fil-
tering using the pval and log
2
FC thresholds respec-
tively. Results from the previous analyses were fur-
ther elaborated to compute the sensitivity and speci-
ficity values associated with the adoption of the differ-
ent methodologies and filtering approaches. Table 2
reports on this data. Specifically, the different rows of
Table 2 show respectively DESeq2, edgeR, DESeq2-
edgeR combined approach and geneEX results. As
geneEx - A Novel Tool to Assess Differential Expression from Gene and Exon Sequencing Data
179

Table 2: Sensitivity/Specificity levels of DESeq2, edgeR, DESeq2-edgeR and geneEX .
pvalue
DESeq2,edgeR
<0.05 log2FC
DESeq2,edgeR
<0.5 pvalue
DESeq2,edgeR,DEX Seq
<0.05 log2FC
DESeq2,edgeR,DEX Seq
<0.5
Sensitivity Specificity Sensitivity Specificity Sensitivity Specificity Sensitivity Specificity
DESeq2 93.5% 29.8% 90.2% 57.8% - - - -
edgeR 94.0% 31.6% 90.7% 60 % - - - -
edger ∩ DESeq2 93.1% 38.7% 89.9% 63.1% - - - -
geneEX - - - - 22.2% 88 % 84.0% 68.4%
previously discussed in terms of TPs, FPs, FNs and
TNs, the adoption of a single tool working at the gene-
expression level did not ensure satisfactory speci-
ficity. With values ranging from 29.8% to 60% de-
pending on the adopted filtering threshold (rows 1 and
2 in Table 2 ). The specificity slightly increased when
adopting a DESeq2-edgeR combined approach, still
maintaining a high sensitivity level. With reference
to geneEX (row 4 of Table 2), its adoption accounted
for an increase in the specificity levels compared to all
the other tested methods and independently from the
adopted filtering measure. However, it is worth not-
ing that the higher improvement was achieved when
filtering according to the pval (specificity equal to
88%). However, this strongly impacted on the sen-
sitivity, reduced to the 22%. A higher sensitivity level
(84%) was instead ensured when filtering according
to the log
2
FC measure while preserving a specificity
level (68.4%) at least 5% higher than that provided by
the compared methodologies. These analyses proved
i) the importance of filtering stages based on exon
analysis to improve specificity and ii) the importance
of an accurate choice of the statistical measure to be
adopted during the different filtering steps. Moreover,
we would like to underline that, as already observed
in the context of ERCC sequence study, the number
of genes investigated in this analysis is very reduced,
due to the lack of datasets coming with a full experi-
mental validation. As consequence, even the number
of FP predictions from the different tools is not so
high. However, in normal experiments FPs are nu-
merous, making impossible the wet-lab validation of
all the results (that comprise both TPs and FPs) from
in-silico analysis and calling for ad-hoc procedures to
shrink the number of candidates focusing on the most
reliable ones.
4 CONCLUSIONS
In this paper, we propose a novel methodology and
tool, namely geneEX, for DEG identification. ge-
neEX integrates the results provided by three widely
adopted bioinformatics algorithms for DEG analysis.
Specifically, it uses DESeq2 and edgeR tools to iden-
tify highly reliable DEG, whereas DEXSeq to further
prioritise these candidates based on exon expression
data. Moreover, by interrogating BioMart database,
geneEX annotates each candidate gene with a series
of information that can be exploited by biologists or
clinicians to focus on the most significant predictions.
Thus, geneEX provides users with a reduced list of
statistically meaningful and highly reliable DEGs to
be deeply investigated by wet-lab experiments. ge-
neEX is highly customizable and easy to use. Users
can trigger its run by specifying both the statistical
measures and the relative values to be adopted dur-
ing DESeq2, edgeR and DEXSeq filtering stages. We
assessed the need for a novel DEG analysis tool on
the ERCC sequences from SEQC dataset. Indeed, this
analysis allowed to highlight the limits of DEG analy-
sis tools, which output lists are generally plagued with
huge amounts of FP predictions, overwhelming wet-
lab validation possibilities. Conversely, geneEX per-
formance was evaluated on four replicates of UHRR
and HBRR datasets, proving the advantages in terms
of specificity increase associated with its adoption.
REFERENCES
Anders, S., Pyl, P. T., and Huber, W. (2015). Htseqa python
framework to work with high-throughput sequencing
data. Bioinformatics, 31(2):166–169.
Canales, R. D., Luo, Y., Willey, J. C., Austermiller, B., Bar-
bacioru, C. C., Boysen, C., Hunkapiller, K., Jensen,
R. V., Knight, C. R., Lee, K. Y., et al. (2006). Eval-
uation of dna microarray results with quantitative
gene expression platforms. Nature biotechnology,
24(9):1115.
Consortium, E. P. et al. (2004). The encode (encyclope-
dia of dna elements) project. Science, 306(5696):636–
640.
DeLuca, D. S., Levin, J. Z., Sivachenko, A., Fennell, T.,
Nazaire, M.-D., Williams, C., Reich, M., Winckler,
W., and Getz, G. (2012). Rna-seqc: Rna-seq metrics
for quality control and process optimization. Bioinfor-
matics, 28(11):1530–1532.
Durinck, S., Moreau, Y., Kasprzyk, A., Davis, S., De Moor,
B., Brazma, A., and Huber, W. (2005). Biomart
and bioconductor: a powerful link between biological
databases and microarray data analysis. Bioinformat-
ics, 21(16):3439–3440.
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B.,
Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y.,
Gentry, J., et al. (2004). Bioconductor: open software
BIOINFORMATICS 2018 - 9th International Conference on Bioinformatics Models, Methods and Algorithms
180

development for computational biology and bioinfor-
matics. Genome biology, 5(10):R80.
Hardcastle, T. J. and Kelly, K. A. (2010). bayseq: empir-
ical bayesian methods for identifying differential ex-
pression in sequence count data. BMC bioinformatics,
11(1):422.
Huber, W., Carey, V. J., Gentleman, R., Anders, S., Carlson,
M., Carvalho, B. S., Bravo, H. C., Davis, S., Gatto, L.,
Girke, T., et al. (2015). Orchestrating high-throughput
genomic analysis with bioconductor. Nature methods,
12(2):115–121.
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley,
R., and Salzberg, S. L. (2013). Tophat2: accurate
alignment of transcriptomes in the presence of inser-
tions, deletions and gene fusions. Genome biology,
14(4):R36.
Lee, C.-Y., Chiu, Y.-C., Wang, L.-B., Kuo, Y.-L., Chuang,
E. Y., Lai, L.-C., and Tsai, M.-H. (2013). Common ap-
plications of next-generation sequencing technologies
in genomic research. Translational Cancer Research,
2(1):33–45.
Li, J., Witten, D. M., Johnstone, I. M., and Tibshirani, R.
(2012). Normalization, testing, and false discovery
rate estimation for rna-sequencing data. Biostatistics,
13(3):523–538.
Li, Y., Rao, X., Mattox, W. W., Amos, C. I., and Liu, B.
(2015). Rna-seq analysis of differential splice junc-
tion usage and intron retentions by dexseq. PloS one,
10(9):e0136653.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated
estimation of fold change and dispersion for rna-seq
data with deseq2. Genome biology, 15(12):550.
Rajkumar, A. P., Qvist, P., Lazarus, R., Lescai, F., Ju, J.,
Nyegaard, M., Mors, O., Børglum, A. D., Li, Q., and
Christensen, J. H. (2015). Experimental validation of
methods for differential gene expression analysis and
sample pooling in rna-seq. BMC genomics, 16(1):548.
Rapaport, F., Khanin, R., Liang, Y., Pirun, M., Krek, A.,
Zumbo, P., Mason, C. E., Socci, N. D., and Betel, D.
(2013). Comprehensive evaluation of differential gene
expression analysis methods for rna-seq data. Genome
biology, 14(9):3158.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010).
edger: a bioconductor package for differential expres-
sion analysis of digital gene expression data. Bioin-
formatics, 26(1):139–140.
Shi, L., Campbell, G., Jones, W., Campagne, F., Wen, Z.,
Walker, S., Su, Z., and Chu, T. Goodsaid, 373 fm,
pusztai, l., et al.(2010). the microarray quality control
(maqc)-ii study of common practices 374 for the de-
velopment and validation of microarray-based predic-
tive models. Nature Biotechnology, 375:28.
Shi, L., Reid, L., Jones, W., Shippy, R., Warrington, J.,
Baker, S., Collins, P., and de Longueville, F. (2006).
Es et al. kawasaki, and maqc consortium. the microar-
ray quality control (maqc) project shows inter-and in-
traplatform reproducibility of gene expression mea-
surements. Nat Biotechnol, 24:1151–61.
Smedley, D., Haider, S., Durinck, S., Pandini, L., Provero,
P., Allen, J., Arnaiz, O., Awedh, M. H., Baldock,
R., Barbiera, G., et al. (2015). The biomart com-
munity portal: an innovative alternative to large, cen-
tralized data repositories. Nucleic acids research,
43(W1):W589–W598.
Smyth, G. K. (2005). Limma: linear models for microar-
ray data. In Bioinformatics and computational biology
solutions using R and Bioconductor, pages 397–420.
Springer.
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L.,
Rinn, J. L., and Pachter, L. (2013). Differential analy-
sis of gene regulation at transcript resolution with rna-
seq. Nature biotechnology, 31(1):46–53.
Young, M. D., McCarthy, D. J., Wakefield, M. J., Smyth,
G. K., Oshlack, A., and Robinson, M. D. (2012).
Differential expression for rna sequencing (rna-seq)
data: mapping, summarization, statistical analysis,
and experimental design. In Bioinformatics for High
Throughput Sequencing, pages 169–190. Springer.
geneEx - A Novel Tool to Assess Differential Expression from Gene and Exon Sequencing Data
181
