
Spot Detection in Microscopy Images using Convolutional Neural
Network with Sliding-Window Approach
Matsilele Mabaso
1
, Daniel Withey
1
and Bhekisipho Twala
2
1
MDS(MIAS), Council for Scientific and Industrial Research, Pretoria, South Africa
2
Department of Electrical and Mining Engineering, University of South Africa, Pretoria, South Africa
Keywords: Microscopy Images, Convolutional Neural Network, Spot Detection.
Abstract: Robust spot detection in microscopy image analysis serves as a critical prerequisite in many biomedical
applications. Various approaches that automatically detect spots have been proposed to improve the analysis
of biological images. In this paper, we propose an approach based on Convolutional Neural Network (conv-
net) that automatically detects spots using sliding-window approach. In this framework, a supervised CNN
is trained to identify spots in image patches. Then, a sliding window is applied on testing images containing
multiple spots where each window is sent to a CNN classifier to check if it contains a spot or not. This gives
results for multiple windows which are then post-processed to remove overlaps by overlap suppression. The
proposed approach was compared to two other popular conv-nets namely, GoogleNet and AlexNet using
two types of synthetic images. The experimental results indicate that the proposed methodology provides
fast spot detection with precision, recall and F_score values that are comparable with the other state-of-the-
art pre-trained conv-nets methods. This demonstrates that, rather than training a conv-net from scratch, fine-
tuned pre-trained conv-net models can be used for the task of spot detection.
1 INTRODUCTION
Object recognition in images has been a major
research area in computer vision that arises in many
real-world applications, such as surveillance (Varga
& Szirányi, 2016), robotics (Wang, et al., 2016),
biology (Li, et al., 2014) and etc. The main goals of
this area are: Firstly, determining what kinds of
objects are present in the image (classification) and,
secondly, the location of these objects in the image
(localization). Knowing which objects are present in
a given image, computing their locations should be
easier; alternatively, knowing where to look,
recognizing the objects should be easier. In other
words, it is important to think of these two tasks
jointly. A lot of existing state-of-the-art object
classification methods does not compute the object
location information.
In this work, we focus on detection of spots in
microscopy images, as shown in Figure 1, but the
methodology can be applied in other applications.
The ability to accurately detect spots is of significant
interests for biomedical researchers as it plays a
significant step for further analysis. A Number of
procedures in biology and medicine require spot
Figure 1: A sample of real fluorescence image with bright
particles obtained using confocal microscopy.
detection and counting, for example, an individual’s
health can be deduced based on the number of red
and white blood cells. Spot detection is interested in
finding all instances of spots in a given image. There
exist several challenges faced by spot detection.
Among them are noise and inhomogeneity which
exist in the background. Besides all these challenges
a lot of applications in bioimage analysis such as
spot tracking (Genovesio, et al., 2006), require high
Mabaso, M., Withey, D. and Twala, B.
Spot Detection in Microscopy Images using Convolutional Neural Network with Sliding-Window Approach.
DOI: 10.5220/0006724200670074
In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018) - Volume 2: BIOIMAGING, pages 67-74
ISBN: 978-989-758-278-3
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
67

performance and reliable detection results which
increases the need for efficiency.
Over the past years, researchers have developed
various methods for the detection of spots in
microscopy images, examples include Wavelets
(Olivo-Marin, 2002), Mathematical morphology
(Kimori, et al., 2010). A detailed review of some of
these methods can be found in (Smal, et al., 2010).
Smal et al. (Smal, et al., 2010) categorized spot
detection methods into ‘supervised’ and
‘unsupervised’ methods. Supervised methods are
machine learning methods which require ground
truth and labeled data for training. Examples of these
methods include Adaptive boosting, Fisher
discriminant analysis. Smal et al. (Smal, et al., 2010)
claimed that these techniques have better detection
performance in the image with low signal-to-noise
ratio (SNR). Unsupervised methods refer to methods
which do not require training. Recent development
in machine learning, namely deep learning has
demonstrated remarkable performance within the
task of image classification.
The convolutional neural network (conv-net) is
one of the popular and effective deep learning
techniques which based on the ImageNet
classification completion which took place 2012,
managed to bring down the error rate by half on the
classification problem. According to He et al. (He, et
al., 2015) a well-trained deep conv-net architecture
can famously perform better than humans in
identifying objects in images. The conv-nets have
since been adopted to various applications in
computer vision community (Noh, et al., 2015) and
medical image analysis (Tajbakhsh, et al., 2016).
Several different conv-nets architectures have since
been developed since 2012, AlexNet (Krizhevsky, et
al., 2012), VGGNet (Simonyan & Zisserman, 2014),
ResNet (He, et al., 2015) and GoogLeNet (Szegedy,
et al., 2015) among others. Despite the range of
their applications in different fields, conv-nets have
only introduced lately to analyze biological data, and
recent works indicate that conv-nets have significant
potential in addressing the needs of a biologist in
analyzing data (Van Valen, et al., 2016).
To our knowledge, there exist no conv-net
architecture for the detection of spots in microscopy
images. As such this work introduces an approach
for the detection of spots based on conv-net and a
sliding window approach. The sliding window is
based on the idea of sliding a box around an image
and classify each image crop inside a box (contains a
spot or not).
This paper is organized as follows: Section 2
describes the methodology used in the study, while
Section 3 presents the results and finally, Section 4
concludes the paper.
2 MATERIAL AND METHOD
2.1 Methodology
2.1.1 Convolutional Neural Network
(Conv-Net)
A convolutional neural network (conv-net) is a
composition of sequence of layers (
……
) that
maps an input vector to an output vector , i.e.,
(1)
where
is the weight and bias vector for the
layer
and
is determined to perform one of the
following: a) convolution with a bank of kernels; b)
spatial pooling; and c) non-linear activation. For any
given training datasets
, we can
estimate the weights,
by solving the
optimization problem:
(2)
Where is defined as the loss function. The
numerical optimization of equation (2) is often
performed via backpropagation and stochastic
gradient descent methods (Ruder, 2017).
2.1.2 Problem Formulation
Given a set of labeled training images, grayscale
image patches defined as
, for in range
1 to with dimensionality for each
image patch. The idea is to train a conv-net to
predict if patch,
contains a spot or not. Image
patches with a full spot contained in the image are
labelled as positive, otherwise negative.
2.1.3 Proposed Convnet
Generally, conv-nets include some of the following
types of layers:
a) Convolution layers, these layers are the
basis of the conv-net architecture and
perform the main computations of the
network including training and firing of
BIOIMAGING 2018 - 5th International Conference on Bioimaging
68
neurons. They work by convolving a kernel
of given size across an input image and
compute the response function over at each
location of the filter.
b) Pooling or down-sampling layers. These
layers are usually put after each conv layer
and reduce the size of the input image for
the next conv layer. It works by sliding a
window and takes the maximum value from
the values within a window at a given
location.
c) Fully connected layers: These layers have
all connection from all neurons in the
previous layer to all output. The main
purpose of the fully connected layer is to
use features form convolutional and
pooling layers for classification of the input
image to various classes. They are typically
used as the last layer in a conv-net, with the
output having one element per class label.
Given the above building blocks, we propose conv-
net architecture for spot detection, named detectSpot
as shown in Table 1. The proposed conv-net consists
of 5 layers (3 convolution layers and 2 fully
connected layers) with learnable weights. We
employ a Rectified Liner Unit (ReLu) (Nair &
Hinton, 2010) activation function for the first four
layers and a softmax for the last layer.
We apply dropout with probability of 0.5 for the
first two fully connected layers (FC). The weights
were initialized using truncated random normal.
Cross-entropy loss was minimized using Adam
optimization with the initial learning rate of 0.001.
Table 1: Proposed conv-net architecture.
Layer
Kernel size,
stride
Output
Input
Conv
ReLu
Max-Pool
Conv
ReLU
Max-Pool
Conv
ReLu
Max-Pool
FC
ReLu+Dropout
FC
ReLu+Dropout
FC
Softmax
2.1.4 Sliding-Window
The procedure adopted for detecting all spots
positions in an image is based on sliding-window
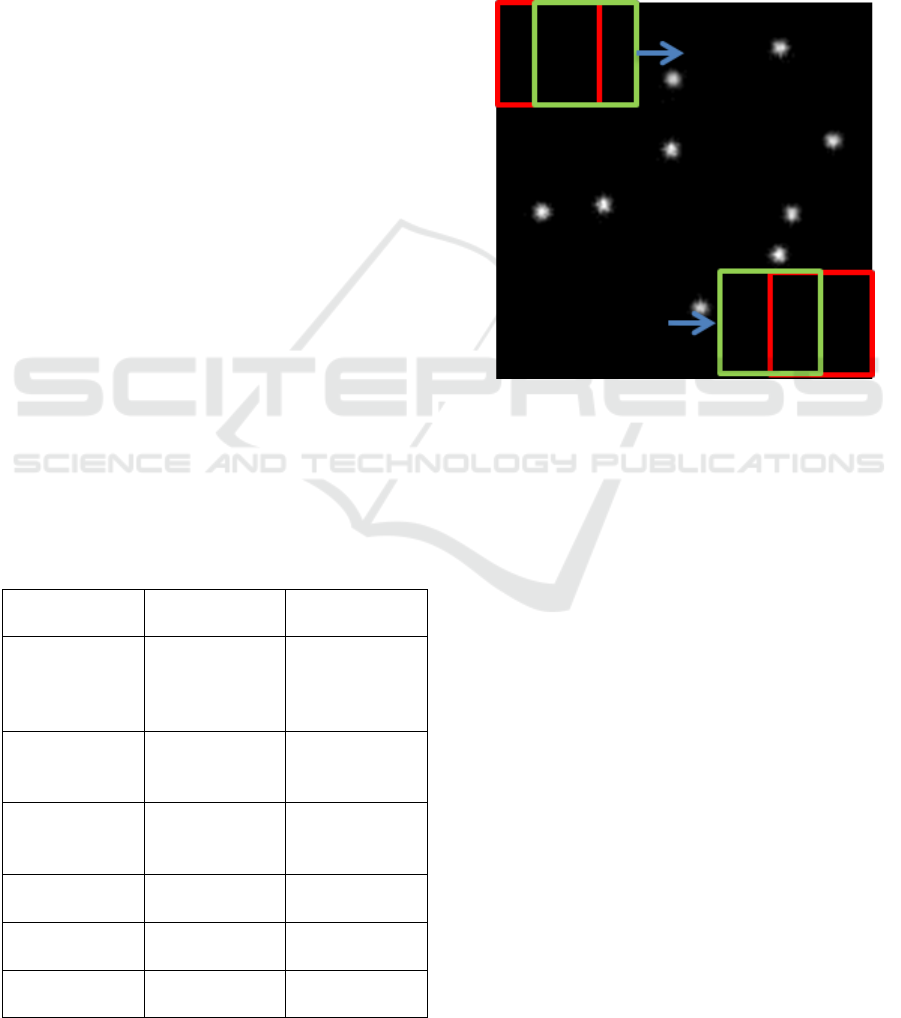
technique. Sliding-window is a technique of sliding
a rectangular window across an image from top to
bottom and left to right as illustrated by red and
green rectangles in Figure 2. This is done in order to
analyze subpart of the image and extract some
information.
Figure 2: Illustration of sliding-window approach.
2.1.5 Dataset
Synthetic image patches sampled from a synthetic
image of size were used for training a
proposed conv-net. Each image patch was of size
pixels. Positive patches were identified as
those which contain a center of a spot and negative
patches are those which do not contain a spot. We
noted that the number of negative patches is usually
disproportionally large compared to the number of
positive patches. This was caused by the fact that
most of each image does not contain spots. Two
measures were then proposed to make training and
validation set more balanced. Firstly, we randomly
discarded negative patches so that the is 50* the
number of positive patches. Secondly, we rotated
each positive patch giving 4 extra positive patches.
A total of 21300 patches created from images with a
signal to noise ratio (SNR) of (20, 10, 5, 2, 1). A
total of 21300 The 21300 image patches were
divided as follows:
• 80% for training
• 20% evaluation
Spot Detection in Microscopy Images using Convolutional Neural Network with Sliding-Window Approach
69

2.1.6 Implementation and Training
To implement and tune a proposed conv-net we used
TFLearn (Damien, 2016). TFLearn is a tensorflow
(Abadi, et al., 2015) wrapper which allows simple
implementation and training of deep learning
models. The network was learned using Adam
(Kingma & Ba, 2015) based optimization algorithm.
Training was carried out on a Linux machine with
16GB RAM and Nvidia GTX680 running TFLearn
(v0.3) and tensorflow (v1.3.0) with.
2.2 Detection of Spots in Test Images
Once the proposed conv-net architecture, deepSpot
is trained it is able to classify an image patch as
containing a spot or not. Figure 3 illustrates the
entire pipeline for the detection of spots. In order to
detect all spots in a complete image, we scan
through an image using a window of size
which is then passed onto a deepSpot and select
those with the highest probability of containing
spot. At each iteration, the extracted sub-window is
Figure 3: The proposed architecture for spot detection in microscopy images.
BIOIMAGING 2018 - 5th International Conference on Bioimaging
70

Figure 4: Examples of synthetic images used for testing with approximately 50 spots per image. (a) Type A, and (b-c)
Type B.
passed onto a classifier to compute a score S, which
defines whether a spot is contained in the sub-
window. Then, if S is bigger than the set threshold T,
the correspond-ding sub-window is considered to
contain spot.
Then, the sub-windows classified as containing
spots are subject to further processing to get spot
centroid
and bounding circles indicating the
location of spots in an image. There are two main
important parameters for our proposed sliding-
window approach, window-size
and stride.
These parameters influence both speed and detection
rate. This approach can only detect spots with fixed
size but it can be extended to spots with different
sizes by introducing image pyramids.
Using a small stride, e.g. stride = 1, will result in
multiple detections of the same spot at slightly
different positions. To overcome this issue, we
group all nearby detections so that every spot is
detected once by using overlap suppression (OS)
approach. The OS method works by grouping all
overlapping detections and suppresses the ones with
lowest scores. This will result in discarding all
overlapping detections.
2.2.1 Using Pre-trained Models
2.2.1.1 Pre-trained Models
The proposed detectSpot model was compared to
two other state-of-the-art conv-nets models, namely,
AlexNet and GoogleNet.
AlexNet: This conv-net was developed by
Krizhevsky et al. (Krizhevsky, et al., 2012) and
successfully applied to large-scale image recognition
and won the ImageNet ILSVRC-2012 challenge.
The model consisted of 8 layers (5 convolutional
layers and three fully connected layers).
GoogleNet: This conv-net was a winner for
ImageNet ILSVRC-2014 proposed by Szegedy et al.
(Szegedy, et al., 2015) from Google. This network
has 12X fewer parameters compared to AlexNet yet
deeper (22 layers). The main contribution of
GoogleNet is the introduction of inception module.
2.3 Synthetic Datasets and Evaluation
Criteria
2.3.1 Synthetic Test Datasets
We generated two types of synthetic datasets (Type
A and Type B) containing spots using a framework
proposed in (Mabaso, et al., 2016) in order to
demonstrate the effectiveness of the proposed
detectSpot model as shown in Figure 4. Each
synthetic image contains 50 spots cluttered on the
background of size pixels. The dataset
was corrupted by white noise. The following signal
to noise ratios (SNR) levels was explored {10, 8, 6,
4, 2, 1} where the spot intensity was 20 gray levels.
The signal to noise ratio is defined as of spot
intensity,
, divided by the noise standard
deviation,
,
(1)
The spot positions were randomized using Icy-
plugin (Chenouard, 2015) to mimic the kinds of
properties in real microscopy images. MATLAB
was used to add spots and the OMERO.matlab-5.2.6
toolbox (Anon., 2016) was used to read and save
images.
2.3.2 Evaluation Criteria
Three state-of-the-art architecture The criteria used
for evaluation is based on computing Precision and
Recall. TP, FP, and FN. The precision, recall, and
is three important measures which are
reported in machine learning research in determining
Spot Detection in Microscopy Images using Convolutional Neural Network with Sliding-Window Approach
71

Figure 5: F_score vs SNR curves for all three conv-nets methods applied to two kinds of synthetic images (a) Synthetic
type A, and (b-c) Synthetic type B.
the performance of the classifier. Precision and
recall are defined in terms of a number of true
positives (TP), false positives (FP) and false
negatives (FN):
(relevant spots detected)
(3)
(spots detected) (4)
(5)
A good detection method should have the value of
approaching one.
3 RESULTS
The trained conv-nets models were each applied to
two types of synthetic images described in Section
2.3.1 as shown in Figure 4 with a signal-to-noise
ratio (SNR) in range {10, 8, 6, 4, 2, 1}. Table 2 -
Table 4 indicates the results for all three conv-nets,
detectSpot, GoogleNet and AlexNet for each of the
test sets. The results were averaged for all SNR’s.
The performance of each method measured using
precision, recall, and F_score. The fair comparison
was achieved by re-training three other conv-nets on
the same datasets. Table 2 indicates that in terms of
Table 2: Evaluation metrics calculated on sythetic images
for three classifiers.
Model
Precision
Recall
GoogleNet
0.833
0.751
0.784
AlexNet
0.842
0.703
0.758
detectSpot
0.836
0.740
0.782
Table 3: Evaluation metrics calculated on realistic
synthetic data. Background 1.
Method
Precision
Recall
GoogleNet
0.717
0.585
0.633
AlexNet
0.443
0.365
0.397
detectSpot
0.803
0.614
0.675
Table 4: Evaluation metrics calculated on realistic
synthetic data. Background 2.
Model
Precision
Recall
GoogleNet
0.733
0.699
0.708
AlexNet
0.567
0.476
0.502
detectSpot
0.780
0.675
0.721
average values, the difference in
performance for GoogleNet and deepSpot is small
compared to AlexNet method. The recall rates are
higher for GoogleNet in Table 2 and Table 4. This
indicates that the method was able to correctly detect
true spots compared to other methods while AlexNet
method has a higher precision. Higher precision
indicate that the method detected less false spots in
comparison to others. However, it shows in Table 3
BIOIMAGING 2018 - 5th International Conference on Bioimaging
72

Figure 6: Results of applying the proposed conv-nets methods on a synthetic image data. Detected spots by each method
are showed in red circles.(a) Original synthetic image. (b) Spots detected by our approach, detectSpot. (c) Detected spots
using using GoogleNet. (d) Detected spots with AlexNet.
and Table 4 that GoogleNet and AlexNet reported
low values for precision compared to detectSpot.
Fig. 5 shows the behavior of each method at all
different signal-to-noise ratios. It can be noted from
the figure that the performance of GoogleNet and
detectSpot is comparable similar for Fig. 5 (a) at
SNR = 10, 8, 2, 4 while AlexNet has higher
at SNR = 1 on Type A images and drops
on Type B images. In Type B synthetic images as
shown in Figure 5(b-c) it indicates that has slightly
higher values for all SNRs. However, the difference
in performance of detectSpot and GoogleNet is
relatively small.
Figure 6 illustrate the performance of each
method on Type A synthetic images with SNR = 10.
4 CONCLUSIONS
Spot detection is an important step towards the
analysis of microscopy images. Over the years,
different approaches have been developed that on
segmentation to perform spot detection.
In this study, we have presented an automated
approach for the detection and counting of spots in
microscopy images, termed detectSpot. The
proposed approach is based on a convolutional
neural network with a sliding-window based
approach to detect multiple spots in images. The
comparative experiments demonstrated that the
GoogleNet and detectSpot methods achieved
comparable performance compared to the AlexNet
method. We also have shown that rather training a
convnet from scratch, knowledge transfer from
natural images to microscopy images is possible. A
fine-tuned pre-trained conv-net can give results
which are comparable to fully trained conv-net.
ACKNOWLEDGEMENTS
This work was carried out in financial support from
the Council for Scientific and Industrial Research
(CSIR) and the Electrical and Electronic
Engineering Department at the University of
Johannesburg.
REFERENCES
Abadi, M. et al., 2015. TensorFlow: Large-scale machine
learning on heterogeneous systems. s.l.:12th USENIX
Symposium on Operating Systems Design and
Implementation.
Anon., 2016. The open microscopy environment. [Online]
Available at: http://www.openmicroscopy.org/site/
support/omero5.2/developers/Matlab.html [Accessed
15 November 2016].
Chenouard, N., 2015. Particle tracking benchmark
generator. [Online] Available at: http://icy.bioimage
Spot Detection in Microscopy Images using Convolutional Neural Network with Sliding-Window Approach
73

analysis.org/plugin/Particle_tracking_benchmark_gen
erator [Accessed 1 November 2016].
Damien, A., 2016. TFLearn. s.l.:GitHub.
Genovesio, A. et al., 2006. Multiple particle tracking in
3d+t microscopy: Method and application to the
tracking of endocytosed quantum dots. IEEE Trans.
Image Process., 15(5), pp. 1062-1070.
He, K., Zhang, X., Ren, S. & Sun, J., 2015. Deep residual
learning for image recognition. s.l., s.n.
He, K., Zhang, X., Ren, S. & Sun, J., 2015. Deep residual
learning for image recognition. s.l., s.n., pp. 770-778.
Kimori, Y., Baba, N. & Morone, N., 2010. Extended
morphological processing: a practical method for
automatic spot detection of biological markers from
microscopic images. BMC Bioinformatics, 11(373),
pp. 1-13.
Kingma, D. P. & Ba, J. L., 2015. Adam: A method for
stochastic optimization. San Diego, s.n.
Krizhevsky, A., Sutskever, I. & Hinton, G. E., 2012.
Imagenet classication with deep convolutional neural
networks. s.l., s.n., pp. 1-9.
Li, R. et al., 2014. Deep learning based imaging data
completion for improved brain disease diagnosis.
Quebec City, s.n.
Mabaso, M., Withey, D. & Twala, B., 2016. A framework
for creating realistic synthetic fluorescence
microscopy image sequences. Rome, s.n.
Nair, V. & Hinton, G. E., 2010. Rectified Linear Units
Improve Restricted Boltzmann Machines. s.l., s.n., pp.
807-814.
Noh, H., Hong, S. & Han, B., 2015. Learning deconvolu-
tion network for semantic segmentation. s.l., s.n.
Olivo-Marin, J.-C., 2002. Extraction of spots in biological
images using multiscale products. Pattern
Recognition, 35(9), pp. 1989-1996.
Ruder, S., 2017. An overview of gradient descent
optimization algorithms. [Online] Available at:
http://ruder.io/optimizing-gradient-descent/[Accessed
10 October 2017].
Simonyan, K. & Zisserman, A., 2014. Very deep
convolutional networks for large-scale image
recognition. s.l., s.n.
Smal, I., Loog, M., Niessen, W. & Meijering, E., 2010.
Quantitative comparison of spot detection methods in
fluorescence microscopy. IEEE Trans on Medical
Imaging, 29(2), pp. 282-301.
Szegedy, C. et al., 2015. Going Deeper with Convolutions.
Boston, s.n.
Tajbakhsh, N. et al., 2016. Convolutional Neural
Networks for Medical Image Analysis: Full Training
or Fine Tuning?. IEEE Transactions on Medical
Imaging, May, 35(5), pp. 1299-1312.
Van Valen, D. A. et al., 2016. Deep learning automates the
quantitative analysis of individual cells in live-cell
imaging experiments. PLoS Comput Biol, November,
12(11), pp. 1-24.
Varga, D. & Szirányi, T., 2016. Detecting pedestrians in
surveillance videos based on convolutional neural
network and motion. Budapest, Hungary, s.n., pp.
2161-2165.
Wang, Z., Li, Z., Wang, B. & Liu, H., 2016. Robot grasp
detection using multimodal deep convolutional neural
networks. Advances in Mechanical Engineering,
August, 8(9), pp. 1-12.
BIOIMAGING 2018 - 5th International Conference on Bioimaging
74
