
A Cloud-aware Autonomous Workflow Engine and Its Application to
Gene Regulatory Networks Inference
Arnaud Bonnaffoux
1,2
, Eddy Caron
3
, Hadrien Croubois
3
and Olivier Gandrillon
1
1
Univ. Lyon, ENS de Lyon, Univ. Claude Bernard, CNRS UMR 5239, INSERM U1210, Lyon, France
2
Cosmo Tech, Lyon, France
3
Univ. Lyon, ENS de Lyon, Inria, CNRS, Universit
´
e Claude-Bernard Lyon 1, Lyon, France
Keywords:
Auto-scaling, Resource Management, Workflow, Cloud, Scientific Applications, HPC.
Abstract:
With the recent development of commercial Cloud offers, Cloud solutions are today the obvious solution for
many computing use-cases. However, high performance scientific computing is still among the few domains
where Cloud still raises more issues than it solves. Notably, combining the workflow representation of com-
plex scientific applications with the dynamic allocation of resources in a Cloud environment is still a major
challenge. In the meantime, users with monolithic applications are facing challenges when trying to move
from classical HPC hardware to elastic platforms. In this paper, we present the structure of an autonomous
workflow manager dedicated to IaaS-based Clouds (Infrastructure as a Service) with DaaS storage services
(Data as a Service). The solution proposed in this paper fully handles the execution of multiple workflows on
a dynamically allocated shared platform. As a proof of concept we validate our solution through a biologic
application with the WASABI workflow.
1 INTRODUCTION
Scientists in fields like biology and physics tend to
rely more and more on High Performance Computing
(HPC) resources both to perform large scale simula-
tions and to analysis the huge amount of data pro-
duced by said simulations as well as other experi-
ments. For example, new generation DNA sequencers
can now produce a large amount of data, in the ter-
abyte range, at a very low cost. Analyzing these re-
quired the development of computing tools for large
scale sequence alignment, which relies upon HPC re-
sources (Das et al., 2017; Yu et al., 2017). Sim-
ilarly, the reconstruction of Gene Regulatory Net-
works (GRNs) from high-throughput experimental
data comes with a high computational cost, leading
to the development of parallel algorithms (Xiao et al.,
2015; Zheng et al., 2016; Lee et al., 2014).
However, accessibility to these HPC resources is
limited and Cloud-based platforms have emerged as
good solution for people with these use-cases that
might not have access to large computing infrastruc-
tures. Using the virtual resources offered by Cloud
providers, anyone can build its own computing plat-
form without having to bear the initial investment cost
and the necessary maintenance that comes with own-
ing the hardware. However, while HPC applications
are moving toward the Cloud, the deployment mecha-
nisms used are generally trying to replicate the exist-
ing paradigm rather than using the full elasticity the
Cloud has to offer.
The approach most commonly used is to de-
ploy Cloud instances such to have a platform simi-
lar to what users are familiar with, and use the batch
scheduling mechanisms they are familiar with. While
this approach requires minimal changes, the tools
used were designed for a fixed platform, and do not
benefit from the dynamicity of Cloud solutions. We,
on the other hand, believe that using the dynamicity of
Cloud infrastructures to modify the platform deploy-
ment in real time can help users achieve better perfor-
mances at a lower cost. Managing such deployment is
a complex task, which requires constant awareness of
the platform and of the workload. In order to achieve
that, we need the (re)deployment mechanisms to work
autonomously. Rather than asking the user to dive
into the details of the platform deployment, we have
to build a solution that releases them from all interac-
tions with this deployment process.
Unlike task placement, which is a well-studied is-
sue, little work deals with the automation of Cloud
platform deployment.
Bonnaffoux, A., Caron, E., Croubois, H. and Gandrillon, O.
A Cloud-aware Autonomous Workflow Engine and Its Application to Gene Regulatory Networks Inference.
DOI: 10.5220/0006772805090516
In Proceedings of the 8th International Conference on Cloud Computing and Services Science (CLOSER 2018), pages 509-516
ISBN: 978-989-758-295-0
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
509

Our goal in this paper is to evaluate the efficiency
of our framework, which contains mechanisms that
automate the deployment of Cloud ressources into a
self adapting, shared, computing platform, as well as
scheduling scientific applications on top of it. As a
proof of concept we validate our solution through a
biologic application with the WASABI workflow. We
provide a tool that contribute to the convergence of
HPC applications and Cloud resources, thus provid-
ing easy access to HPC resources to all users.
2 RELATED WORK
Many scientific and industrial applications from vari-
ous disciplines are structured as workflows (Bharathi
et al., 2008). A workflow can be seen as a structured
set of operations which, given an input data set, pro-
duce the expected result. For a long time, the devel-
opment of complex middleware with workflow en-
gine (Couvares et al., 2007; Deelman et al., 2005;
Caron et al., 2010) automated workflow management.
Infrastructure as a Service (IaaS) Clouds raised a lot
of interest recently thanks to an elastic resource allo-
cation and pay-as-you-go billing model. A Cloud user
can adapt the execution environment to the needs of
their application on a virtually infinite supply of re-
sources. While the elasticity provided by IaaS Clouds
gives way to more dynamic application models, it also
raises new issues from a scheduling point of view. An
execution now corresponds to a certain budget, which
imposes certain constraints on the scheduling process.
Solutions to dynamically-scaled Cloud computing
instances exist. For example in (Mao et al., 2010)
the solution is based on deadline and budget informa-
tion. In (Kailasam et al., 2010) the solution deals with
Cloud bursting. Another autonomous auto-scaling
controller, which maintains the optimal number of re-
sources and responds efficiently to workload varia-
tions based on the stream of measurements from the
system, is introduced in (Londo
˜
no-Pel
´
aez and Florez-
Samur, 2013). This paper shows the benefits of an
auto-scaling solution for Cloud deployments. How-
ever, these solutions do not handle workflow appli-
cations. Likewise in (Nikravesh et al., 2015), au-
thors gave a suitable prediction technique based on
the performance pattern, which led to more accurate
prediction results. Unfortunately, all these papers fail
to consider the delays resulting from communications
between the different tasks of a workflow.
In (Mao and Humphrey, 2013), authors show
the benefit of auto-scaling to deal with unpredicted
workflow jobs. They also show that scheduling-first
and scaling-first algorithms have different advantages
over each other within different budget ranges. The
auto-scaling mechanism is introduced as a promising
research direction for future work.
Nevertheless, some solutions provide an au-
tonomous workflow engine. In (Heinis et al., 2005),
altering the cluster configuration helps the authors
build an autonomous controller that responds to work-
load variations. Authors introduce a mechanism of
self-healing that reacts to changes in the cluster con-
figuration. This solution shows some benefits for
cluster architecture but does not work well for Cloud
architecture. Workflow engines for Cloud environ-
ments dealing with dynamic scalable runtimes are
given in (Pandey et al., 2012).
A comparative evaluation of several auto-scaling
algorithms is given in (Ilyushkin et al., 2017). How-
ever, the policies discussed in the review solely focus
on auto-scaling, thus missing on issues like data local-
ity and tasks clustering. Our approach is different in
that it considers a more complex issue where work-
flow optimizations can induce cycles in their clus-
tered representation. This representation ask for more
complex task placement and demand analysis mecha-
nisms. Last but not least, our approach differs in that
parts of the resource manager and of the scheduler are
decentralized and rely on the nodes to take decisions
by themselves in order to improve scalability.
3 INFRASTRUCTURE
Our objective in this paper is to describe and evaluate
the architecture of a middleware that uses resources
from Cloud providers to build a computing infrastruc-
ture for the execution of scientific workflows.
3.1 IaaS Cloud Platforms
Among the many offers Cloud providers propose,
IaaS (Infrastructure as a Service) is arguably one of
the most versatile. Through the use of virtualization
technologies, it allows anyone to get access to remote
resources and use them just as if they owned their own
server. These resources are seen as virtual machines
and they can run any system the user needs. This en-
ables anyone to build their own computing infrastruc-
ture without the initial investment cost or the burden
of maintenance that comes with owning hardware.
The main advantage of Cloud solutions, which ex-
plains their development over the past few years, is
the versatility and dynamicity of these solutions. Not
only can a user deploy a custom platform in just min-
utes, but the deployment can be modified to match any
change in the workload. As such, users only have to
CLOSER 2018 - 8th International Conference on Cloud Computing and Services Science
510
pay for what is really needed, and not bear the cost of
platform when not in use.
3.2 The Allocation Problem
It is easy to deploy a large platform using current
Cloud technologies. However, knowing how many
resources are really needed is a completely different
problem. There might be many options, each one re-
sulting in different performances and costs. While
cost per unit of time is easy to compute, estimating
performances and tasks completion time is extremely
difficult. Consequently, the total deployment cost can
also be hard to predict as it depends on completion
time.
A common practice is to distribute the budget
along a specific duration and get as many resources
as one can afford during this period. This simple ap-
proach, which tries to maximize performances within
a given cost constraint, has major drawbacks. Not
only is the user committing all his budget, but there
are no warranties to have enough resources during
peak hours, and resources are very likely to be wasted
during off-peak hours.
A more elegant and efficient solution would be to
modify the deployment in real time so that it matches
the needs. Estimating the needs is a very complex
task, and adjustment should be performed 24/7, which
would require too many actions to be realistically per-
formed by a human. For it to be efficient, we need
this process to be fully autonomous and not require
any human input.
3.3 An Autonomic Solution
In order to automate the deployment of the platform,
we have to design a control loop that could drive
the allocation mechanisms. According to the MAPE-
K (Kephart and Chess, 2003) model, such a loop re-
quires 4 features: (1) Monitoring the platform, both in
terms of platform status and workload; (2) Analyzing
the needs in term of platform allocation; (3) Planning
actions to modify platform allocation towards what is
required; (4) Executing the plan.
Such design is already part of common schedulers
and is used to control the placement of the interde-
pendent tasks in a workflow. However, unlike depen-
dency control, the issue of platform deployment con-
trol is complex and still unsolved. This is this issue
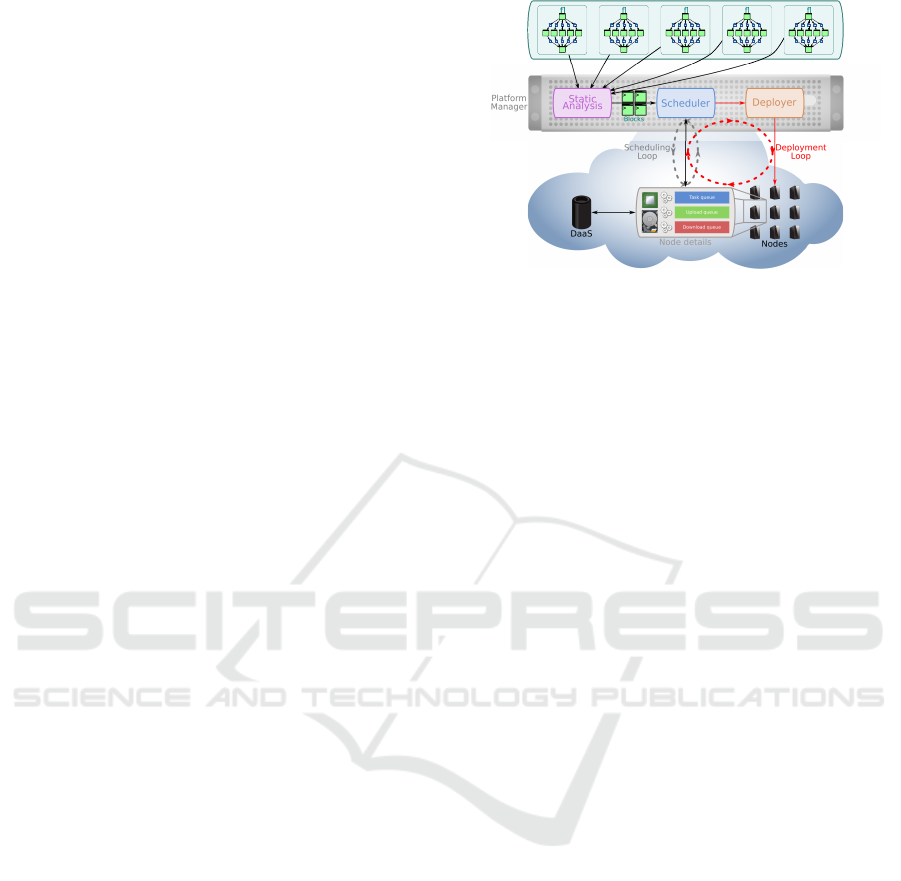
that our approach (Figure 1) tries to solve in the con-
text of homogeneous IaaS Clouds.
In addition to the traditional mechanisms used to
control the task flow (scheduling loop), we added fea-
tures to the existing agents, as well as new agents,
>
>>> > >
>
>
>>> > >
>
>
>>> > >
>
>
>>> > >
>
>
>>> > >
>
Cloud
Users
Work ow 1 Work ow 2 Work ow 3 Work ow 5Work ow 4
Figure 1: Outlines of our middleware with autonomous
platform deployment capabilities.
to implement a deployment control loop. More pre-
cisely, we built these such that two distinct mecha-
nisms work in coordination to achieve the automation
of both up-scaling and down-scaling.
Data Locality. Data locality is a major concern in
the scheduling of workflows. While parallelization is
expected to reduce the completion time of workflows,
data transfer can produce undesirable side effect that
reduces the overall performances.
In our previous work (Caron and Croubois, 2017)
we discussed the possibility to solve this issue using
an off-line scheduling mechanism. The static anal-
ysis step described in this paper left us with blocks
of tasks that already handle the issue of data locality
and can therefore be scheduled without requiring the
scheduler to solve this optimization issues at runtime.
Down-scaling Control. The down-scaling is de-
centralized and controlled by the nodes themselves.
Each node has its own work queue, and requests work
from the scheduler when it has free resources in term
of CPU (ability to start new work) or Down-link (abil-
ity to prefetch data). When a node requests work,
the scheduler is to provide it with a block that can
be executed on this node without breaking the dead-
line constraints. However, if the platform is oversized,
the nodes will end up executing all the tasks up to
the point where the scheduler cannot provide work to
the requesting node. In addition to requesting work,
when a node work queue is empty, the node starts a
suicide timer. Once this timer is initiated, and if no
work has been received by then, the node will auto-
matically deallocate itself before the beginning of the
next billing hour.
This mechanism helps reduce the dimension of
oversized platforms during off-peak hours thus en-
abling a more efficient platform deployment that de-
creases infrastructure cost.
A Cloud-aware Autonomous Workflow Engine and Its Application to Gene Regulatory Networks Inference
511

Up-scaling Control. Unlike the down-scaling, the
up-scaling mechanism is not decentralized, and re-
quires a global knowledge of the platform. It relies
on a new agent, the deployer. When the workload
changes, for example with the addition of new jobs
by the users, the scheduler will inform the deployer of
these changes, and provide it with a description of the
work queues and nodes status. This monitoring of the
platform means that the deployer is able to use simple
list scheduling algorithms to simulate jobs placement,
analyze it, and plan for the deployment of new nodes
if required.
If new nodes have to be deployed, the deployer
will do so according to the last computed deployment
schedule. This schedule can be overridden by new,
updated, runs of the analysis and planning steps. This
control mechanism ensures new nodes are deployed
when required, such that QoS is achieved (deadlines
are meet).
4 GENE REGULATORY
NETWORKS INFERENCE
4.1 WASABI
Gene Regulatory Networks (GRN) play an important
role in many biological processes, such as cell dif-
ferentiation, and their identification has raised great
expectations for understanding cell behaviors. Many
computational GRN inference approaches are based
on bulk expression data, and they face common issues
such as data scarcity, high dimensionality or popu-
lation blurring (Chai et al., 2014). We believe that
recent high-throughput single cell expression data
(see (Pina et al., 2012)) acquired in time-series will
allow to overcome these issues and give access to
causality, instead of “simple” correlations, to dissect
gene interactions. Causality is very important for
mechanistic model inference and biological relevance
because it enables the emergence of cellular decision-
making. Emergent properties of a mechanistic model
of a GRN should then match with multi-scale (molec-
ular/cellular) and multi-level (single cell/population)
observations.
The WASABI (WAves Analysis Based Inference)
framework is based upon the idea of an iterative infer-
ence method. This will allow to adopt a divide-and-
conquer type of approach, where the complexity of
the problem is broken down to a “one gene at a time”
much simpler problem, which can be parallelized.
The whole process can be decomposed along the
following steps:
1. Gene Ordering. We order all the 94 genes from
time-series single cell gene expression data from
chicken erythrocyte progenitors acquired during
their differentiation process (Richard et al., 2016).
These data have been analyzed using a stochastic
mechanistic model of gene expression, the Ran-
dom Telegraph model (Peccoud and Ycart, 1995).
2. Iterative inference. For each step of the inference
process a new gene is added to a set of GRN can-
didates inferred in the previous iterations. It cre-
ates new extended GRN candidates to be assessed,
though all possible combinations with the previ-
ously conserved networks. For each new network,
its behavior will be simulated using a recently de-
scribed mathematical formalism (Herbach et al.,
2017) and the resulting gene expression values
will be compared to the experimental one. Should
the fit between the two genes expression be ac-
ceptable, the networks will be kept and carried
to the next step. If, on the contrary, the network
should be too different, then it will be pruned and
will not participate in future attempts.
At the beginning of the process, one would expect
(and an initial assessment confirmed) that the num-
ber of suitable networks will sharply increase. In this
growth phase, an efficient parallelization will be of
essence. In a second phase, one expects that most of
the ”bad” networks will have accumulated so many
errors that they will diverge from the experimental re-
ality, and it will in the end result in the generation of
a manageable amount of networks.
The use of Design of Experiment ap-
proaches (Kreutz and Timmer, 2009) should
finally help us to get to the most probable network.
4.2 WASABI Workflow Description
WASABI, as an application, is composed of many
steps, each one divided into many instances of the
same elements. The control flow, which links these
many elements into the complete application, can ex-
press this iterative structure through a DAG of suc-
cessive fork-join patterns. The WASABI workflow
contains 9 fork-join steps of width 5, 6, 36, 252,
1000, 1000, 1000, 1000, 1000. This adds up to a to-
tal 5309 tasks (including synchronization tasks) and
10598 control flow dependencies. Total runtime for
all these tasks is 4250.1 hours.
All these informations, as well as details about in-
dividual tasks, such as runtime) were extracted from
traces of the previous runs. This allowed us to build
a descriptions of WASABI as a workflow. While our
middleware is not based on the Pegasus engine, for
the sake of clarity, our inputs file are compliant with
CLOSER 2018 - 8th International Conference on Cloud Computing and Services Science
512

the well-known Pegasus workflow syntax. It is this
workflow that we will be using to compare our de-
ployment solution to existing approaches.
5 EVALUATION
5.1 Comparison to Fixed Deployment
for a Single Workflow Execution
In order to estimate the efficiency of our deployment,
we will compare it to a more traditional “fixed deploy-
ment” method. In a fixed deployment, the user books
a given amount of resources which are then used by a
batch scheduler. Whenever a task is ready to run and a
node is available, the task will be placed on the node
to be computed. It is clear that, with this approach,
the more resources there are, the faster the workflows
will be executed, up to the point where the sequen-
tial dimension of the workflow prevents the user from
achieving any more parallelism. Still, having more re-
sources also means that more cpu time is wasted dur-
ing the synchronization parts of the workload. Once
all computation is done, the user has to release the
resources.
Assessing total cost and runtime of a workflow for
a specific deployment is a hazardous operation which
requires trial and error. Simulation can help with
this process, but it requires knowledge of the plat-
form specifications and time, which users non famil-
iar with computer science might not have. What our
framework offers is a platform where users can spec-
ify their workflows and the required wall time for each
of them. The platform is then automatically deployed
in order to meet the expected QoS while achieving the
lowest cost possible.
The results in this section have be achieved
through simulation. Details about the WASABI work-
flow were extracted from traces of runs on existing
HPC structures (IN2P3). Estimation of the deploy-
ment cost for those simulations were obtained assum-
ing their deployment on Amazon EC2 instances of
similar performance. The down-scaling mechanism
was tunned to match Amazon EC2 billing policy.
Our first results show platforms performance and
cost for deploying a single WASABI workflow. This
workflow contains 5309 tasks for a total of 4250 core-
hours. Ideally, we would like to pay only for these
4250 hours of computation, but the billing policy of
Cloud providers is such that we will also be charged
for some unused time when we cannot perfectly use
the hours allocated to each node. For example, a node
used to compute a task that lasts 1 hour and 48 min-
utes will be billed for 2 hours, and we are thus wast-
ing 12 minutes of CPU time. The critical path in this
workflow is about 17 hours, meaning that no matter
how many resources we have, we will not be able to
get results faster than that using the type of node con-
sidered here.
For this experiment, we run simulations with fixed
platforms of sizes varying from 100 to 400 nodes.
This gives us a base line of what current approaches
achieve. The results in Table. 1b and Figure 1a show
a Pareto front which comes from the conflict between
two contradictory objectives: maximizing platform
performance and minimizing deployment cost. This
confirms the idea that there is not ideal value for the
size of a fixed platform. Selecting the size of a fixed
platform results in a choice between performance and
cost on this non-optimal front.
However, the dynamicity offered by the deploy-
ment control loop implemented in our framework
leads to better results. By efficiently allocating and
deallocating nodes we manage to free ourselves from
this Pareto efficiency and we achieve good results in
term of deployment cost even when facing tight QoS
constraints.
5.2 Multi-tenant/Multi-workflows
Deployment: Example of a Small
Lab Using WASABI
In the previous section, we saw how our approach can
efficiently deploy a computing platform to execute a
single instance of the WASABI workflow. However,
this context still requires quite many human interven-
tions as the platform was dedicated to this workflow.
This means that in order to execute their workflow, the
user first has to deploy the platform manager. Even
worse, in the case of a fixed allocation, the user has
to deploy the fixed platform, selecting the number of
nodes they want and to be there at the end of the run
to shut down the platform.
We believe that the platform should be
maintenance-free, and users should be able to
submit workflows to an already existing platform
manager. This perpetually running tool would be the
entry where any user can submit their workflows. The
platform would be handled automatically, allocating
new nodes when needed, sharing nodes between
workflows of different users and shutting nodes down
when they are no longer necessary.
Sharing a computing platform today, using the
fixed deployment approach and a batch scheduler, is
simple but very inefficient. In addition to the waste
we could already witness when running a single work-
flow, we also have to consider all the wasted resources
A Cloud-aware Autonomous Workflow Engine and Its Application to Gene Regulatory Networks Inference
513
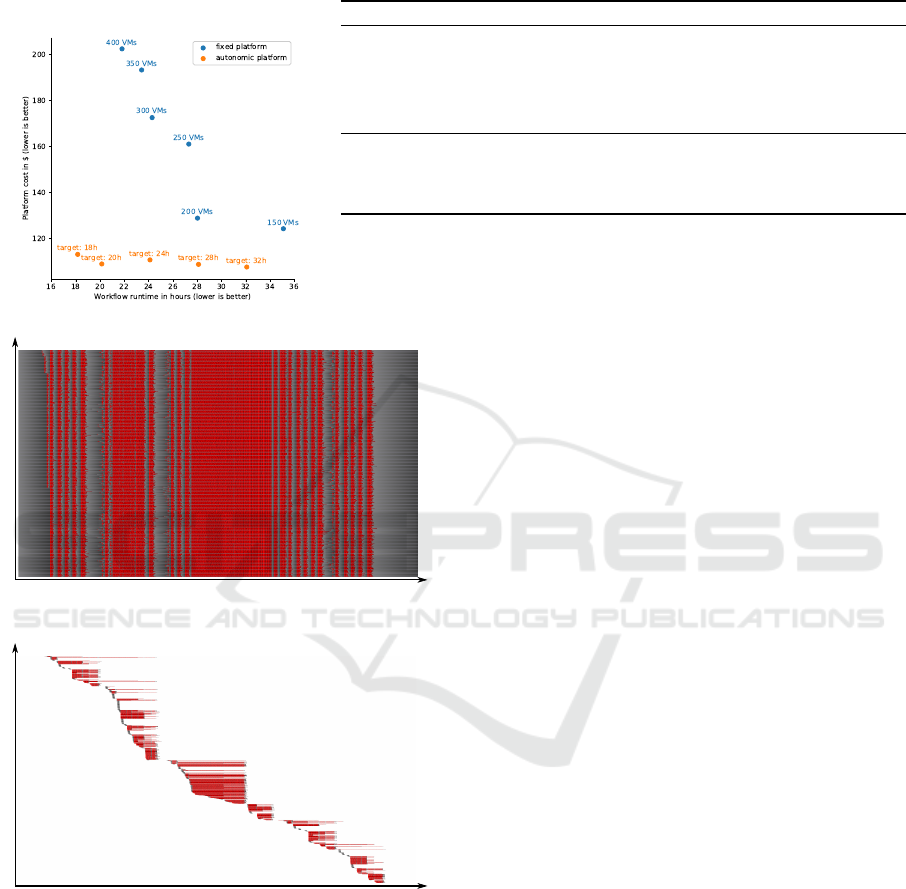
Table 1: Details for different runs of the WASABI workflow on various platforms. The workflow contains 5309 tasks for a
total of 4250.1 core-hours.
(a) Cost/Makespan front (b) Statistics
Platform #VMs Walltime VM duration Total usage Cost Efficiency
Fixed (100) 100 47:10:51 48h 4800 core-hours $110.400 88.54%
Fixed (150) 150 35:04:51 36h 5400 core-hours $124.200 78.71%
Fixed (200) 200 28:00:49 28h 5600 core-hours $128.800 75.89%
Fixed (250) 250 27:17:42 28h 7000 core-hours $161.000 60.72%
Fixed (300) 300 24:16:44 25h 7500 core-hours $172.500 56.67%
Fixed (350) 350 23:24:56 24h 8400 core-hours $193.200 50.60%
Fixed (400) 400 21:48:05 22h 8800 core-hours $202.400 48.30%
Autonomous (18h) 851 18:09:14 Variable 4915 core-hours $113.045 86.47%
Autonomous (20h) 711 20:08:05 Variable 4734 core-hours $108.882 89.78%
Autonomous (24h) 653 24:06:05 Variable 4811 core-hours $110.653 88.34%
Autonomous (28h) 676 28:05:46 Variable 4726 core-hours $108.698 89.93%
Autonomous (32h) 663 32:03:59 Variable 4676 core-hours $107.548 90.89%
Note: Cost were computed assuming Amazon EC2 t2.small instances for an on-demand price of $0.023/hour.
Time
Nodes
(a) Fixed platform
Time
Nodes
(b) Autonomous platform
Figure 2: Gantt diagram of VMs during the multi-tenants
execution of WASABI workflows. 10 workflows are exe-
cuted over a one week period, for a total of 42501.0 core-
hours of computation. For each VM (line) computation time
is shown in red and idle time in grey. Figure 2a shows re-
sults for fixed deployment of 500 VMs running for the fun
weeks while Figure 2b shows result for an autonomous de-
ployment that spawned 4698 VMs over the week, some of
which only ran for a single hour.
which are allocated during off-peak hours.
We simulated such a context at the scale of a small
laboratory. In this example, we consider a research
team who uses the WASABI application to analyze
their results. Over a one week period, our imagi-
nary lab produces data requiring 10 runs of WASABI.
Researchers in this team submit the data for analy-
sis when available. As such, the submissions are not
distributed evenly throughout the week. In particular,
we assume that all submissions will be performed dur-
ing work days. While no new jobs are submitted dur-
ing the week-end, computation can still be performed
during the week-end. When using the autonomous
approach, they ask the results to be available 24 hours
after submission.
Statistics for this simulated workload is reported
in Table. 2. Gantt diagrams for both deployment
methods are shown in Figure 2. Once again, we see
that the dynamic allocation of node has major advan-
tages over the previous approach.
The most obvious advantage to our method is cost.
Having a platform with 500 t2.small instances run-
ning for a whole week would cost $1932.00, with an
efficiency of 50.60% with this particular workload.
For the same work, our approach would reduce de-
ployment cost by 44.57%.
The second advantage to our method is the ease of
maintenance and the efficiency of the elastic deploy-
ment.
Some might argue that the fixed platform does not
have the right size, and using 500 nodes is an em-
pirical choice, and they would be right. However, as
discussed previously, choosing which platform size to
use is not an obvious choice. While having a 500
nodes platform leads to wasted resources, it is also not
enough to ensure that sufficient resources are avail-
CLOSER 2018 - 8th International Conference on Cloud Computing and Services Science
514

Table 2: Platforms statistics for the multi-tenants execution of WASABI workflows. 10 workflows are executed over a 1 week
period, for a total of 42501.0 core-hours of computation.
Platform #VMs Fastest run Slowest run Total VM usage Cost Efficiency
Fixed 500 20:12:36 33:58:52 84000 core-hours $1932.00 50.60%
Autonomous 4698 23:50:03 24:07:12 46563 core-hours $1070.95 91.28%
Note: Cost were computed assuming Amazon EC2 t2.small instances for an on-demand price of $0.023/hour.
able to all users. During rush periods, workflows are
fighting for resources. This is visible around the mid-
dle of Figure 2a when all nodes are busy. During
this period, no resources are wasted but work does not
progress as users would expect, causing some work-
flows to finish with delays. While users would require
the workflows to be computed within 24 hours, some
took over 33 hours due to the limited number of re-
sources available.
Unlike the fixed platform approach, the au-
tonomous deployment allocates resources when
needed which limits waste during off-peak hours and
guarantees that the user will have the results they ex-
pect with minimal delays. This is visible in Figure
2b. The burst in the workload causes the nodes to
be allocated and deallocated in blocks. When many
resources are required, the platform manager deploys
many nodes. Once the work has been done, nodes
start to suffer work shortage and start deallocating.
We can see that the burst that caused the fixed plat-
form to saturate is here triggering the allocation of
many nodes. These nodes execute tasks from all ac-
tive workflows and contribute to meeting the QoS
users expect. In our example, this elastic allocation
process helped limiting delays to at most 7 minutes
12 second.
We also notice that the efficiency achieved by our
method in the multi-tenants context is slightly bet-
ter than in a single workflow context. On a fixed
platform, having multiple workflows causes conflicts
and decreases the QoS. However, with our elastic de-
ployment manager, having multiple workflows on the
same platform is a good thing as the workflows can
share nodes to maximize their use and reduce waste.
QoS is maintained by the allocation of new resources
when required.
Last but not least, the autonomous deployment
makes the results availability more predictable. This
is more comfortable for users who might require these
results for specific deadlines or just to be part of a
broader pipeline. With a fixed platform, users are af-
fected by each other’s submissions which might lead
to frustration and conflicts.
6 CONCLUSION
In this paper, we presented the outlines of a fully au-
tonomous platform manager. We also described the
WASABI application, and how it can benefit from this
platform management. Comparing this approach to
more traditional ones, we showed that not only do we
achieve the QoS required by the users through dead-
lines, but we also achieve substantial savings in de-
ployment cost.
So far this approach is restricted to homogeneous
infrastructures, and further work is required to make
this solution more versatile. Other optimization ob-
jectives, such as minimizing a workflow execution
time with a constrained budget, also require some
work. Still, our multi-tenants example showed that
using our platform to deploy existing scientific work-
loads can result in cost savings of 40% while avoiding
the need for human intervention in the management of
the platform.
In conjunction with previous work on the static
clustering of workflows for DaaS-based Cloud execu-
tion, we now have a comprehensive solution. While
the modular nature of this framework means the com-
ponents could be improved, the current state of this
solution is already effective and we hope to apply it
soon in the context of an easy-to-use, all-in-one solu-
tion for scientific and industrial users.
REFERENCES
Bharathi, S., Chervenak, A., Deelman, E., Mehta, G., Su,
M.-H., and Vahi, K. (2008). Characterization of sci-
entific workflows. In SC’08 Workshop: The 3rd Work-
shop on Workflows in Support of Large-scale Science
(WORKS08) web site, Austin, TX. ACM/IEEE.
Caron, E. and Croubois, H. (2017). Communication aware
task placement for workflow scheduling on daas-
based cloud. In Workshop PDCO 2017. Parallel / Dis-
tributed Computing and Optimization, Orlando, FL.
USA. In conjunction with IPDPS 2017, The 31st IEEE
International Parallel & Distributed Processing Sym-
posium.
A Cloud-aware Autonomous Workflow Engine and Its Application to Gene Regulatory Networks Inference
515

Caron, E., Desprez, F., Glatard, T., Ketan, M., Montag-
nat, J., and Reimert, D. (2010). Workflow-based
comparison of two distributed computing infrastruc-
tures. In Workflows in Support of Large-Scale Science
(WORKS10), New Orleans. In Conjunction with Su-
percomputing 10 (SC’10), IEEE. hal-00677820.
Chai, L. E., Loh, S. K., Low, S. T., Mohamad, M. S., Deris,
S., and Zakaria, Z. (2014). A review on the compu-
tational approaches for gene regulatory network con-
struction. Comput Biol Med, 48:55–65.
Couvares, P., Kosar, T., Roy, A., Weber, J., and Wenger, K.
(2007). Workflow Management in Condor. In Taylor,
I., Deelman, E., Gannon, D., and Shields, M., editors,
Workflows for e-Science, pages 357–375. Springer.
Das, A. K., Koppa, P. K., Goswami, S., Platania, R., and
Park, S. J. (2017). Large-scale parallel genome assem-
bler over cloud computing environment. J Bioinform
Comput Biol, 15(3):1740003.
Deelman, E., Singh, G., Su, M.-H., Blythe, J., Gil, Y.,
Kesselman, C., Mehta, G., Vahi, K., Berriman, G. B.,
Good, J., Laity, A., Jacob, J., and Katz, D. (2005). Pe-
gasus: a Framework for Mapping Complex Scientific
Workflows onto Distributed Systems. Scientific Pro-
gramming Journal, 13(3):219–237.
Heinis, T., Pautasso, C., and Alonso, G. (2005). Design
and evaluation of an autonomic workflow engine. In
Autonomic Computing, 2005. ICAC 2005. Proceed-
ings. Second International Conference on, pages 27–
38. IEEE.
Herbach, U., Bonnaffoux, A., Espinasse, T., and Gan-
drillon, O. (2017). Inferring gene regulatory net-
works from single-cell data: a mechanistic approach.
https://arxiv.org/abs/1705.03407.
Ilyushkin, A., Ali-Eldin, A., Herbst, N., Papadopoulos,
A. V., Ghit, B., Epema, D., and Iosup, A. (2017).
An experimental performance evaluation of autoscal-
ing policies for complex workflows. In Binder, W.,
Cortellessa, V., Koziolek, A., Smirni, E., and Poess,
M., editors, ICPE, pages 75–86. ACM.
Kailasam, S., Gnanasambandam, N., Dharanipragada, J.,
and Sharma, N. (2010). Optimizing service level
agreements for autonomic cloud bursting schedulers.
In Parallel Processing Workshops (ICPPW), 2010
39th International Conference on, pages 285–294.
IEEE.
Kephart, J. O. and Chess, D. M. (2003). The vision of auto-
nomic computing. Computer, 36(1):41–50.
Kreutz, C. and Timmer, J. (2009). Systems biology: exper-
imental design. FEBS J, 276(4):923–42.
Lee, W. P., Hsiao, Y. T., and Hwang, W. C. (2014). De-
signing a parallel evolutionary algorithm for inferring
gene networks on the cloud computing environment.
BMC Syst Biol, 8:5.
Londo
˜
no-Pel
´
aez, J. M. and Florez-Samur, C. A. (2013). An
autonomic auto-scaling controller for cloud based ap-
plications. International Journal of Advanced Com-
puter Science and Applications(IJACSA), 4(9).
Mao, M. and Humphrey, M. (2013). Scaling and schedul-
ing to maximize application performance within bud-
get constraints in cloud workflows. In Parallel & Dis-
tributed Processing (IPDPS), 2013 IEEE 27th Inter-
national Symposium on, pages 67–78. IEEE.
Mao, M., Li, J., and Humphrey, M. (2010). Cloud auto-
scaling with deadline and budget constraints. In Grid
Computing (GRID), 2010 11th IEEE/ACM Interna-
tional Conference on, pages 41–48. IEEE.
Nikravesh, A. Y., Ajila, S. A., and Lung, C.-H. (2015).
Towards an autonomic auto-scaling prediction system
for cloud resource provisioning. In Inverardi, P. and
Schmerl, B. R., editors, 10th IEEE/ACM International
Symposium on Software Engineering for Adaptive and
Self-Managing Systems, SEAMS 2015, Florence, Italy,
May 18-19, 2015, pages 35–45. IEEE Computer Soci-
ety.
Pandey, S., Voorsluys, W., Niu, S., Khandoker, A., and
Buyya, R. (2012). An autonomic cloud environment
for hosting ecg data analysis services. Future Gener-
ation Computer Systems, 28(1):147–154.
Peccoud, J. and Ycart, B. (1995). Markovian modelling of
gene product synthesis. Theoretical population biol-
ogy, 48:222–234.
Pina, C., Fugazza, C., Tipping, A. J., Brown, J., Soneji, S.,
Teles, J., Peterson, C., and Enver, T. (2012). Inferring
rules of lineage commitment in haematopoiesis. Nat
Cell Biol, 14(3):287–94.
Richard, A., Boullu, L., Herbach, U., Bonnafoux, A.,
Morin, V., Vallin, E., Guillemin, A., Papili Gao,
N., Gunawan, R., Cosette, J., Arnaud, O., Kupiec,
J. J., Espinasse, T., Gonin-Giraud, S., and Gandrillon,
O. (2016). Single-cell-based analysis highlights a
surge in cell-to-cell molecular variability preceding
irreversible commitment in a differentiation process.
PLoS Biol, 14(12):e1002585.
Xiao, X., Zhang, W., and Zou, X. (2015). A
new asynchronous parallel algorithm for inferring
large-scale gene regulatory networks. PLoS One,
10(3):e0119294.
Yu, J., Blom, J., Sczyrba, A., and Goesmann, A. (2017).
Rapid protein alignment in the cloud: Hamond com-
bines fast diamond alignments with hadoop paral-
lelism. J Biotechnol.
Zheng, G., Xu, Y., Zhang, X., Liu, Z. P., Wang, Z., Chen,
L., and Zhu, X. G. (2016). Cmip: a software pack-
age capable of reconstructing genome-wide regulatory
networks using gene expression data. BMC Bioinfor-
matics, 17(Suppl 17):535.
CLOSER 2018 - 8th International Conference on Cloud Computing and Services Science
516
