
MATESC: Metadata-Analytic Text Extractor and Section Classifier
for Scientific Publications
Maria F. De La Torre, Carlos A. Aguirre, BreAnn M. Anshutz and William H. Hsu
Department of Computer Science, Kansas State University, 2184 Engineering Hall, Manhattan, KS, U.S.A.
Keywords: Structured Information Extraction, Document Analysis, Text Analytics, Metadata, Classification,
Information.
Abstract: This paper addresses the task of extracting free-text sections from scientific PDF documents, and specifically
the problem of formatting disparity among different publications, by analysing their metadata. For the purpose
of extracting procedural knowledge in the form of recipes from papers, and for the application domain of
nanomaterial synthesis, we present Metadata-Analytic Text and Section Extractor (MATESC), a heuristic
rule-based pattern analysis system for text extraction and section classification from scientific literature.
MATESC extracts text spans and uses metadata features such as spatial layout location, font type, and font
size to create grouped blocks of text and classify them into groups and subgroups based on rules that
characterize specific paper sections. The main purpose of our tool is to facilitate information and semantic
knowledge extraction across different domain topics and journal formats. We measure the accuracy of
MATESC using string matching algorithms to compute alignment costs between each section extracted by
our tool and manually-extracted sections. To test its transferability across domains, we measure its accuracy
on papers that are relevant to the papers that were used to determine our rule-based methodology and also on
random papers crawled from the web. In the future, we will use natural language processing to improve
paragraph grouping and classification.
1 INTRODUCTION
MATESC is a metadata-analytic text extractor and
section classifier that uses metadata features and
heuristics to classify examined text elements that are
extracted from Portable Document Format (PDF)
scientific publications into titled sections and
subsections. Examples of metadata features include
font size, font type, and spatial location of elements
that can in turn be localized using computer vision
and pattern recognition algorithms. MATESC was
designed to be a generalized extractor whose
functionality is transferable across different domain
topics and journal publishers. The purpose of section
classification in our extraction task is to address the
problem of IR-based and knowledge-based question
answering (QA), which requires the extraction of
passages directly from documents, guided by the text
of the user question, to formulate a structured
response (Jurafsky et al., 2009).
Given the potentially enormous amount of text
and information that can be retrieved from a
document, section extraction for QA tasks entails
narrowing down search sections, controlling a user
interface to focus on specific sections and passages of
interest, and reducing costs of extracting answers for
specific predetermined user queries or search-based
QA. In fields such as material science, QA tasks
require the extraction of domain-specific information,
such as recipes for synthesizing a material of interest
(Kim et al., 2017). These are stepwise procedures
consisting of named compounds and operations. Our
goal is to use MATESC to obtain specific text
sections, such as “Materials and Methodology”, that
can be annotated to obtain training data for machine
learning algorithms, resulting in models for natural
language processing such as snippet and passage
extraction, named entity recognition (NER), set
expansion, relationship extraction, chunk parsing,
and semantic role labelling. This in turn allows new
documents to be tagged with mark-up for snippets or
passages, named entities, chemical terms and the
acronyms and synonyms, “verbs” denoting unit
operations or sub-procedures, unknown terms in the
form of noun phrases, and recognizable roles of
recipe ingredients. The end-to-end function of this
Torre, M., Aguirre, C., Anshutz, B. and Hsu, W.
MATESC: Metadata-Analytic Text Extractor and Section Classifier for Scientific Publications.
DOI: 10.5220/0006937702610267
In Proceedings of the 10th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2018) - Volume 1: KDIR, pages 261-267
ISBN: 978-989-758-330-8
Copyright © 2018 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
261

cognitive computing pipeline uses text and
knowledge features to drive a process based on semi-
supervised learning to produce material synthesis
recipes.
This paper presents a rule-based algorithm used to
extract section titles, beyond header information, and
group lines of text in their corresponding paragraphs
while placing those paragraphs in their correct
sequential order. To measure the effectiveness of our
algorithm in section classification and ordering, we
developed a user interface to manually extract section
titles and their content from 300 documents to create
our ground truth. With the purpose of creating a
transferable tool across different domain topics, we
compared efficiency measures between the domain-
topic used to develop MATESC, material synthesis,
and other random domains. Half of the documents
were relevant to material synthesis determined by
field professionals, and the other half were randomly
crawled from the web using the open-source web-
crawling platform, Scrapy (Myers et. al., 2015). The
length of the longest common subsequence (LLCS)
(Paterson et. al., 1994), and the length of the longest
common substring, (LLCSTR) (Crochemore et al.,
2015) were measured to determine similarity,
precision, recall and accuracy between the manually
extracted ground truth and the sections extracted by
MATESC. For ordering of section measurements, we
use different variations of k, which determines
comparison of sections only if they k indices apart.
1.1 Background
QA tasks rely heavily on the amount of information
publicly available in the world wide web (Jurafsky et.
al, 2009). With the tremendous growth of scientific
documents publicly available, the format disparity
across different publishers and domain topics
increases. Although there seems to be a general
guideline for scientific papers, there are various
format differences that bring challenges in handling
this disparity to create a generalized tool. In some
documents, section subtitles are not included, making
it difficult for natural language processing to parse
header data.
To address format disparity challenges, metadata
extraction tools have been developed for specific
entities extraction, specifically headers (e.g. title,
authors, keywords, abstract) and bibliographic data.
Apache PDFBox (Apache, 2018), PDFLib TET
(PDFLib, 2018) and Poppler (Noonburg, 2018)
extract text and attributes of PDF documents. Open-
source header and bibliographic data parsers include
GROBID (Lopez, 2009), ParsCit (Prasad et al., 2018)
and SVMHeaderParse (Han et al. 2003) For table and
figures extraction, PDFFigures (Clark et. al., 2016)
and Tabula (Aristaran et. al., 2013) have been
developed for general academic publications. To
encapsulate all of these various open source tools into
one framework, PDFMEF (Wu et al., 2015) brings
users a customizable and scalable tool to bring the
best capabilities of each tool into one tool. Extraction
of first-page header information is useful for
clustering documents and identifying duplicates,
where a combination of authors and title are assumed
to be unique to each document. For structured recipe
extraction, sections beyond the first page and
bibliographic data are necessary to extract step-like
recipe entities. GROBID has been shown to have
advantages over other methods in first-page and
bibliographic sections (Lipinski et al., 2013). Other
sections, e.g. materials, methodology, results and
discussion, are not fully extracted or classified by the
mentioned tools and are often in the wrong order. For
recipe extraction, sequential order is essential for the
accurate extraction of synthesis steps. In this paper,
we compare the accuracy, precision, and recall (based
on edit distance) of three products of information
extraction: (1) manually extracted ground truth (text
selected and ordered by manual annotation); (2) the
section output of GROBID (Lopez, 2009); and (3) the
output of MATESC.
1.2 Applications
MATESC is the metadata-aware payload extraction
component of a broader project whose long-term goal
is to acquire a corpus of scientific and technical
documents that are restricted to a specific domain and
extract free-text recipes consisting of procedural steps
and entities organized in a sequential form. For our
specific application domain of nanomaterials
synthesis, the documents of interest are academic
papers collected from open-access web sites using a
custom crawler and scraper ensemble. The initial
seeds for the document crawl were provided by the
subject matter expert. The papers to be analyzed by
MATESC are PDF files, from which structured
information such as titles, author lists, keyword lists,
sets of figures with captions, and specific named
sections such as the introduction, background and
related work, experimental method, result data, and
summary and conclusions, are captured. The next
stage of analysis is to extract recipes, which are
sequences of steps that specify materials needed and
methods utilized to produce a nanomaterial. These are
similar in structure and length to cooking recipes.
Steps of a recipe may consist of basic unit operations
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
262

or intermediary multi-step methods that are
composed of more primitive steps. The framework
and algorithms of MATESC itself are not limited to
the domain of nanomaterials alone; there are
applications in many other scientific and technical
fields that require reading large numbers of
documents and would benefit from being able to
filter, rank, and extract structured information by
means of section and passage extraction, followed by
shallow parsing at the sentence level. Examples of
such applications include the medical and legal
domains, where there are large text collections for
specific professional purposes that practitioners
regularly sift through in order to obtain procedural
information.
2 METHODOLOGY
MATESC takes as input text information extracted
using PyMuPDF (Liu et. al, 2018), a tool that
provides metadata features about each character,
including font type, font size and spatial location
relative to each pdf page. The input text is filtered and
cleaned by removing rare Unicode characters and
irrelevant information usually found in the margins of
each document page, using their spatial location.
These include publication identifiers, headers and
footers with page numbers, and watermarks. After the
text is cleaned, our algorithm uses heuristics to merge
each character into its corresponding line, while
considering font and spatial location differences to
differentiate between section titles and section
content. Those lines are then grouped into paragraphs
and ordered, considering single, double and triple-
column documents in reconstructing a sequential
order. Figure 1 shows the workflow of MATESC,
from the input data stage to the output stage, which is
customizable for XML, HTML or JSON output. Each
step is described in detail in the following section.
2.1 Line Assembly
Spatial location is helpful when determining whether a
character belongs to the same line as the previous
word or
to the following line. MATESC uses x, y coordinates,
font type, and font size to merge lines of characters.
If the character is within a specified y range, that
considers subscripts and superscripts, of the previous
character, we append the character to that line.
Moreover, during line assembly, if the next
character’s y range (line height) is overlapping the
current line’s y range, then it is appended to it,
otherwise it is the beginning of a new line. Because
Figure 1: MATESC’s input processing and section
classification.
mathematical and chemical formulae are important
for information extraction in material synthesis,
MATESC checks for subscripts as well; this can be
challenging because the y range can extrude a
MATESC: Metadata-Analytic Text Extractor and Section Classifier for Scientific Publications
263

variable amount above or below a character, causing
the algorithm to assign the subscript or superscript to
a new line. To handle this issue, x coordinates are
considered: if the character is off in the y range but is
in proximity of the previous and next character in
terms of x coordinates, then it is recognized as a
subscript and merged with the line of the preceding
character. Moreover, for each character, font type and
size are considered. If font type or size changes and
the list of those characters are between a certain range
length, then that line is extracted as a subtitle, their
position and metadata features are saved and are later
used for section extraction and ordering.
2.2 Paragraph Assembly
After all characters have been merged into their
corresponding lines, and subtitles have been extracted
based on their metadata features, those lines are
grouped into paragraphs. Here, the x and y
coordinates are considered. If two lines are in an
extremely close range of x coordinates and their
distance in y (vertical distance) is less than the height
of a character, then those two lines are assigned into
the same paragraph. Each paragraph is assigned a
bounding box for which spatial location, and an
associated average font size and type, are calculated.
It is important to pass these metadata features for the
paragraph down the pipeline because these features
will be used for paragraph sequential ordering.
2.3 Paragraph Order
Before we can classify each paragraph into sections,
we must sequentially order all paragraphs. Here, we
must consider the number of columns used in that
particular section, which determines the heuristics
used to order the paragraphs by x or y coordinates
first. We get an idea of the structure of each page by
calculating the ratio between each the x coordinate
length and the length of the page without margins.
This ratio allows us to determine the number of
columns in each page (e.g., single, two-column, and
three-column). Then, depending on the column, we
use different rules for paragraph ordering. If the page
contains a single column, then we simply order by x.
If it consists of two or three columns, we order
separately by y for those paragraphs that are in the
same x range. Those groups are assigned to a column,
and then those columns are ordered by the x
coordinates of their bounding boxes.
2.4 Paragraph Classification
Once all the paragraphs are in the correct order, we
can begin to classify each paragraph onto their
corresponding sections. We use the subtitles extracted
in Section 2.1, and, based on their spatial location, we
assign everything between that section title and the
next one to that section title. Moreover, once all of the
paragraphs have been assigned to a section, we use
the spatial location and page number of each subtitle
to perform an overall sequential ordering of all the
sections. If no subtitles were found, we use column
information to differentiate between abstract and
body. Since it is common for an abstract to be single-
column, while the rest of the paper is two-column.
3 EXPERIMENT DESIGN
3.1 Evaluation Method: Manual
Extraction for Ground Truth
To evaluate the output of MATESC, we manually
extracted sections from 300 papers to obtain a
reference version (the designated ground truth) and
compared this against two automatically-generated
outputs: that of the chemical IE system GROBID and
that of MATESC.
The manual extraction process to produce each
payload, a reference extract in raw unformatted text
form, consists of simple highlighting (copying) of
contiguous sections of text, one column block at a
time. A human annotator must exercise judgement to
make decisions on the extent of a column block and
the ordering of these blocks when pasting them into a
file.
Because MATESC was designed for the purpose
of IE from papers in a specific domain of interest -
nanomaterials synthesis - it is important to test its
generalization quality. To test transferability across
various domain fields and journals, the experimental
corpus was deliberately constructed using 150 papers
known to be relevant to our application domain plus
another 150 random PDFs scraped from the web
using a built-in random file selection function of the
Scrapy web crawling framework (Myers et. al.,
2015).
3.2 Distance Metrics for Text
The evaluation approach consists of computing
distance metrics between reference (ground truth) and
automatic extracts. We use distance metrics for text
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
264
alignment as in common practice in bioinformatics
(Xia, 2007) and payload-extraction approaches to
web page cleaning (Marek et al., 2008, Weninger et
al., 2010).
The overall IE system within which our text
payload extraction task fits is geared towards
capturing all text related to a recipe, and ultimately
extracting a structured representation of that recipe.
The system thus includes a separate pipeline to
extract images, tables, figure captions, chemical and
mathematical formulas. However, the output of
MATESC omits such text snippets, resulting in a
penalty to its score because such omissions would be
scored as deletions from the reference extract.
To account for this issue, we consider two
measures of string comparison: Longest Common
Substring (LCSTR) and Longest Common
Subsequence (LCS). LCSTR finds the longest
substring(s) between two strings, while LCS finds the
longest string that is a shared subsequence between
two strings, allowing for position disparity in
individual words. LCS is thus a more tolerant
measure for standalone algorithms and heuristics
designed to extract separate components of the
payload. This metric is more salient to our task as it
can ignore strings that are passed to an independent
pattern recognition subsystem, rather than penalizing
for their omission.
We use the length of these two resulting strings,
LCSTR and LCS, to compute precision, recall, and
accuracy.
4 RESULTS
4.1 Random Documents
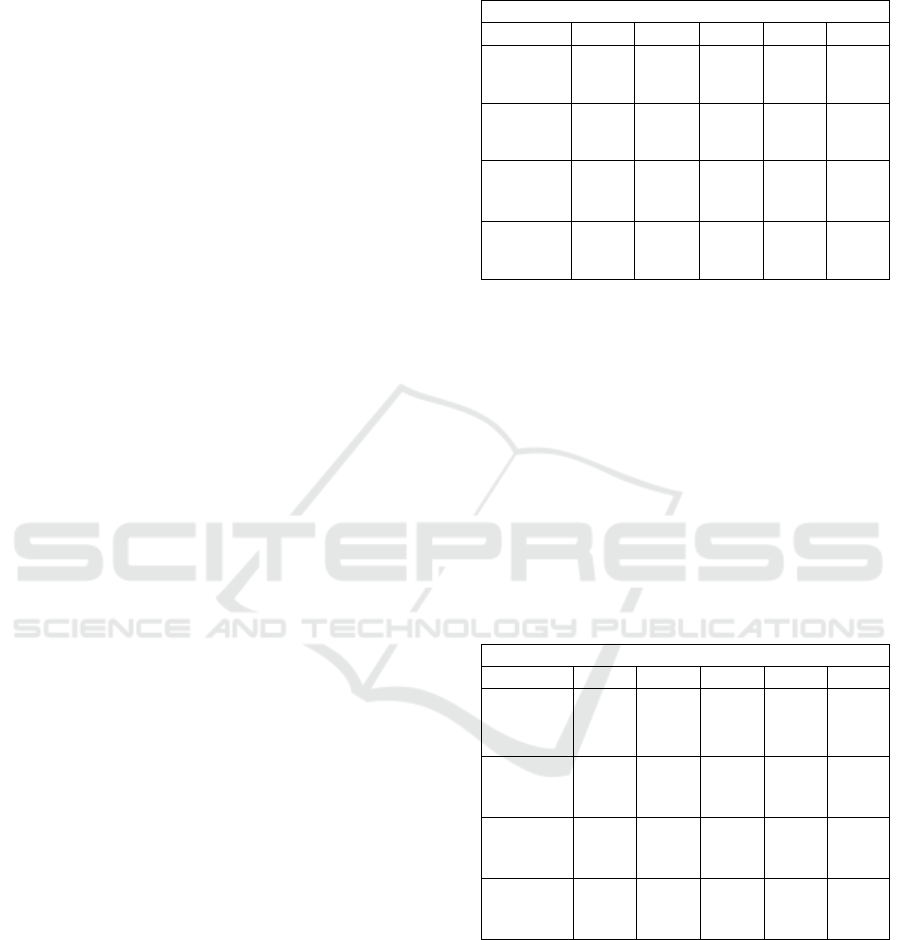
Table 1 shows the average scoring results for all
sections on random papers. Using LCS, the null
hypothesis that GROBID classifies a greater number
of words into their corresponding section than
MATESC is rejected with p < 0.000000122 (1.22 ✕
10-7) at the 95% level of confidence using a paired,
one-tailed t-test on their F1 scores. On the other hand,
using LCSTR the null hypothesis fails to be rejected
with p < 0.09449 at the 95% level of confidence using
a paired, one-tailed t-test on their F1 scores.
Table 1: Precision, Recall, Accuracy and F1 for random
papers across MATESC and GROBID using LCS and
LCSTR.
Random Papers
TPR FPR PPV ACC F1
MATESC
LCSTR
0.119 0.208 0.166 0.681 0.106
MATESC
LCS
0.629 0.109 0.631
0.849
0.573
GROBID
LCSTR
0.095 0.132 0.148 0.716 0.097
GROBID
LCS
0.418 0.066 0.566 0.819 0.437
4.2 Domain-Relevant Documents
For domain-relevant papers, Table 2 shows the
average scoring results for all sections. The null
hypothesis that GROBID classifies a greater number
of words into their corresponding section than
MATESC using LCSTR and LCS is rejected with p <
8.99 ✕ 10
-10
and p < 2.38 x 10
-29
at the 95% level of
confidence using a paired, one-tailed t-test on their F1
scores.
Table 2: Precision, Recall, Accuracy and F1 for relevant
papers across MATESC and GROBID using LCS and
LCSTR.
Domain-Relevant Papers
TPR FPR PPV ACC F1
MATESC
LCSTR
0.133 0.267 0.179 0.601 0.128
MATESC
LCS
0.737 0.082 0.776
0.879
0.723
GROBID
LCSTR
0.087 0.161 0.210 0.631 0.091
GROBID
LCS
0.373 0.063 0.580 0.755 0.392
4.3 Sections
Averages for each individual section are shown on
Table 3, we show precision, recall, accuracy and F1
scores for only general sections (title, authors,
abstract, keywords, methodology, results,
conclusions, acknowledgments, references) using
both LCS and LCSTR for MATESC. While Table 4
shows the results for only random papers are shown
MATESC: Metadata-Analytic Text Extractor and Section Classifier for Scientific Publications
265

given the importance of transferability across
different domains. Other sections that are specific to
each paper are not shown in the table as they cannot
be averaged across documents. For LCS, it is
observed that the precision, recall and F1 score of
GROBID are on average higher than those of
MATESC for title and abstract, and introduction;
while for all other sections, the output of MATESC
scores higher. For LCSTR, the precision, recall and
F1 score of GROBID are on average higher than those
of MATESC for title, abstract, introduction,
methodology and results. These findings are in
keeping with the modular design principle of our
overall IE system including MATESC and the
hypothesis that LCS is a more lenient metric across
the board but also a more salient one for such modular
systems.
5 CONCLUSIONS
5.1 Summary and Interpretation of
Results
As expected, the results for LCS are on average better
than the results for LCSTR across both types of
papers and extractors. For random papers, in the case
of LCSTR, results for GROBID and MATESC are
not statistically different, which can be explained by
the development focus of MATESC on a scientific
domain relevant to those for which GROBID was
designed. However, the LCS score for the output of
MATESC was slightly better than that of GROBID
for random paper; the LCSTR scores for MATESC
were comparable to those of GROBID for relevant
papers and the LCS scores were substantially better,
as expected due to our development focus.
In the case of particular sections, for titles, authors
and reference sections, the output of GROBID is
expected to be more accurate than that of MATESC,
as that is the design focus of GROBID and not of our
system. From the results reported in the preceding
section we infer that GROBID outperforms
MATESC on authors and references because its
output for those more structured sections contain
more information (e.g., university, address, phone
numbers) than our manually extracted authors, which
only contained the first and last name of each author,
similarly with references.
Overall, MATESC performed in average similar
or better than a well-established text extractor such as
GROBID.
5.2 Future Work
Machine learning approaches using as features
computer vision data and text analytics are likely to
improve MATESC heuristics on header and footer
text, figure and table text, and subtitle recognition.
Learning to classify techniques on header and footer
text can increase the FPR on section bodies. Similar
techniques can be used to determine whether text
belongs to the body of a section or if it is part of a
figure or table. Finally, MATESC uses section titles
as section delimiters, therefore a better section title
recognition mechanism can aid in identifying correct-
ly whether a new section begins and where it ends.
For future experimentation, a larger experimental
corpus is needed, and is being developed.
Furthermore, we plan to compare our approach to
other text extraction and section classification
approaches. Another measurement that could help to
draw further insights would be the Levenshtein
Distance (LD) which calculates the edit distance of
two strings considering deletions, insertions and
substitutions. This would give us a penalty score in
which we can compare different extractors (or
versions of our extractor) without the cost of
performing the calculations for both LCS and
LCSTR. This could help in the development task by
decreasing the time of testing.
Finally, the document analysis task presented in
this paper constitutes a key part of a procedural
pipeline for recipe extraction, namely, taking a
clipping of a document. Continuing work on machine
learning explores the use of deep reinforcement
learning for this clipping subtask, to learn extraction
policies and representation. Related tasks of semi-
supervised and transfer learning also arise from the
need to extract sections that have a positive
downstream impact on capture of procedural
knowledge, and ultimately actionable recipes.
ACKNOWLEDGEMENTS
This work was funded by the Laboratory Directed
Research and Development (LDRD) program
at Lawrence Livermore National Laboratory
(16-ERD-019). Lawrence Livermore National
Laboratory is operated by Lawrence Livermore
National Security, LLC, for the U.S. Department of
Energy, National Nuclear Security Administration
under Contract DE-AC52-07NA27344. This is also
supported by the U.S. National Science Foundation
(NSF) under grants CNS-MRI-1429316 and EHR-
DUE-WIDER-1347821.
KDIR 2018 - 10th International Conference on Knowledge Discovery and Information Retrieval
266

REFERENCES
Apache, Apache PDFBox | A Java PDF Library. Available
at: https://pdfbox.apache.org/ [Accessed May 24,
2018a].
Aristaran, M., Tigas, M. and Merrill, J. (2013). Tabula:
Extract Tables from PDFs. [online] Tabula.technology.
Available at: https://tabula.technology/ [Accessed 20
Jul. 2018].
Clark, C. and Divvala, S. (2016). PDFFigures 2.0.
Proceedings of the 16th ACM/IEEE-CS on Joint
Conference on Digital Libraries - JCDL '16.
Crochemore, M. et al., 2015. The longest common
substring problem. Mathematical Structures in
Computer Science, 27(02), pp.277–295.
Han, H. et al., 2003. Automatic document metadata
extraction using support vector machines. Proceedings
of the 2003 Joint Conference on Digital Lib. Available
at: http://dx.doi.org/10.1109/jcdl.2003.1204842.
Jurafsky, D. and James, H.M. (2009) Speech and Language
Processsing.2nd edn. Upper Saddle River, NJ, USA:
Prentice-Hall.
Kim, E. et al., 2017. Materials Synthesis Insights from
Scientific Literature via Text Extraction and Machine
Learning. Chemistry of materials: a publication of the
American Chemical Society, 29(21), pp.9436–9444.
Lipinski, M. et al., 2013. Evaluation of Header Metadata
Extraction Approaches and Tools for Scientific PDF
Documents. Available at: https://books.google.com/
books/about/Evaluation_of_Header_Metadata_Extract
ion.html?hl=&id=j7JCAQAACAAJ.
Liu, R., & McKie, J. X.., PyMuPDF. Available at:
http://pymupdf.readthedocs.io/en/latest/ [Accessed
May 24, 2018].
Lopez, P., 2009. GROBID: Combining Automatic
Bibliographic Data Recognition and Term Extraction
for Scholarship Publications. , pp.473–474.
Marek, M., Pecina, P., & Spousta, M., 2007. Web page
cleaning with conditional random fields. In Fairon, C.,
Naets, H., Kilgarriff, A., & de Schryver, G.-M., eds.
_Building and Exploring Web Corpora (WAC3 - 2007):
Proceedings of the 3rd web as corpus workshop,
incorporating cleaneval_, pp. 155-162.
Myers, D. & McGuffee, J.W., 2015. Choosing Scrapy.
Journal of Computing Sciences in Colleges, 31(1),
pp.83–89.
Noonburg, D., Poppler. Available at: http://poppler.
freedesktop.org [Accessed May 24, 2018].
Paterson, M. & Dančík, V., 1994. Longest common
subsequences. In Mathematical Foundations of
Computer Science 1994. International Symposium on
Mathematical Foundations of Computer Science.
Springer, Berlin, Heidelberg, pp. 127–142.
PDFLib, TET. Available at: http://www.pdflib.com/
products/tet/ [Accessed May 24, 2018b].
Prasad, A., Kaur, M. & Kan, M.-Y., 2018. Neural ParsCit:
a deep learning-based reference string parser.
International Journal on Digital Libraries. Available
at: http://dx.doi.org/10.1007/s00799-018-0242-1.
Weninger, T., Hsu, W. H., & Han, J., 2010. CETR - content
extraction via tag ratios. In Proceedings of the 19th
International World Wide Web Conference (WWW
2010), pp. 971-980. doi:10.1145/1772690.1772789
Wu, J. et al., 2015. PDFMEF. In Proceedings of the
Knowledge Capture Conference on ZZZ - K-CAP 2015.
Available at: http://dx.doi.org/10.1145/2815833.28158
34.
Xia, X., 2007. _Bioinformatics and the Cell: Modern
Computational Approaches in Genomics, Proteomics
and Transcriptomics_, pp. 24-48. New York, NY, USA:
Springer.
MATESC: Metadata-Analytic Text Extractor and Section Classifier for Scientific Publications
267
