
Molecular Docking Study of Lemon (Citrus limon (Linn) Burm. f)
Flavonoid Derivatives Compound in Receptor Cyclooxygenase-1
(COX-1) as Antiplatelet in Ischaemic Stroke Disease
Rizky Arcinthya Rachmania*, Hariyanti, Nurul Rochmah
Faculty of Pharmacy and Science, Universitas Muhammadiyah Prof. DR. HAMKA (UHAMKA)
Islamic Center, Jl. Delima II/IV Perumnas Klender, Jakarta Timur, 13460, Telp. (021) 8611070
Keywords: Citrus limon, COX-1, flavonoid derivatives, antiplatelet, docking.
Abstract: Lemon (Citrus limon (Linn) Burm. f) is a plant that has efficacy as antiplatelet. Flavonoids in lemon
potentially obstruct COX-1 (Cyclooxygenase-1) receptor which has an important role in increasing
thromboxane A2 in the process of ischaemic stroke. This research aims at looking for flavonoids activity
from citrus lemon which is expected to be the antiplatelet drug candidates. The method used in this research
was the molecular docking method using Autodock Vina and Pymol software programs. The results showed
that the value of ΔG binding affinity Aspirin as the standard ligand was -6,5 kcal/mol and lemon flavonoid
derivatives that have the lowest ΔG binding affinity value was on Neohesperidin -15,4 kcal/mol and Rutin -
15,3 kcal/mol. This research shows that Neohesperidin and Rutin in lemon can be used as drug candidate of
antiplatelet in ischaemic stroke disease.
1 INTRODUCTION
Stroke is the third most common disease after heart
disease and cancer. According to the World Health
Organization (WHO), the definition of stroke is a
rapidly growing clinical sign of focal (or global)
brain dysfunction, with symptoms that last for 24
hours or more, can cause death, with no cause other
than vascular. Stroke is characterized by a sudden
loss of blood circulation to the brain area resulting in
a neurological deficit (Gund et al., 2013). In general,
strokes are classified as ischaemic and hemorrhagic.
Ischaemic stroke is an ischaemic brain tissue arising
from a blockage in the cervical vascular blood
vessels or brain tissue hyperfusion by various factors
such as atherothrombosis, embolism, or
hemodynamic instability (Chung and Caplan, 2007).
Stroke is the third leading cause of death in the
United States and Britain after heart disease and
cancer, and the main cause of adult disability. In the
United States, more than 160,000 adult Americans
die of strokes every year. In Europe, around 650,000
people died from strokes. In the United States,
people who report strokes over 65 years (Gund et
al., 2013). According to the latest data from Riset
Kesehatan Dasar in 2013, the number of stroke
patients in Indonesia in 2013 based on the diagnosis
of Health Workers is estimated to be 1,236,825
people (7.0 ‰), whereas based on diagnosis
symptoms are estimated as many as 2,137,941
people (12.1 ‰) (Balitbang, 2013).
Antiplatelet therapy is an important long-term
treatment for all patients at risk of atherothrombosis
such as ischaemic stroke. A comparison of some
antiplatelet drugs statistically indicated a significant
difference in outcomes (Shinohara et al., 2010).
Strong platelet function inhibitors have been
developed in recent years with different drug-action
mechanisms, because when combined the effects are
additive or even synergistic (Ringleb et al., 2011).
These antiplatelet drugs are classified into several
groups based on their mechanism of action, namely
inhibition of prostaglandin synthesis (aspirin),
inhibition of ADP-induced platelet aggregation
(clopidogrel, prasugrel, ticlopidine), and blockade of
the receptor glycoprotein IIb/IIIa in platelets
(abciximab, tirofiban, and eptifibatide) (Ringleb et
al., 2011). However, these drugs can cause serious
side effects for users such as gastrointestinal
bleeding, leukopenia, and thrombocytopenia
(Ringleb et al., 2011). Because of the side effects of
Rachmania, R., Hariyanti, . and Rochmah, N.
Molecular Docking Study of Lemon (Citrus limon (Linn) Burm. f) Flavonoid Derivatives Compound in Receptor Cyclooxygenase-1 (COX-1) as Antiplatelet in Ischaemic Stroke Disease.
DOI: 10.5220/0008238700190025
In Proceedings of the 1st Muhammadiyah International Conference on Health and Pharmaceutical Development (MICH-PhD 2018), pages 19-25
ISBN: 978-989-758-349-0
Copyright
c
2021 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
19

these drugs, herbal medicine is also an option for
patients.
Indonesia has a wealth of herbs, one of which
is lemon (Citrus limon (Linn) Burm F.). One of the
compounds in lemon that is thought to be potential
as an antiplatelet is flavonoids. Based on studies that
have been conducted, lemon plants are known to
have activity as an anticoagulant and antiplatelet in
vitro/in vivo (Riaz et al., 2014). Flavonoids are one
type of antioxidant that can inhibit adhesion,
aggregation and platelet secretion (Retnaningsih et
al. 2007). The ability of flavonoids to inhibit platelet
aggregation is caused by the flavonoid inhibiting the
metabolism of arachidonic acid by cyclooxygenase
enzyme, thus reducing the amount of thromboxane
A2 (TXA2) production and platelet aggregate
production causing blood vessel blockage
(Middleton et al., 2000).
Cyclooxygenase (COX) is a functional enzyme
bound to the membrane acting to catalyze two
important stages in the formation of prostanoid,
cyclooxygenation, and peroxidation reaction. The
cyclooxygenation reaction stage is the stage at which
COX conducts a cyclization process and the addition
of two oxygen molecules to arachidonic acid to form
prostaglandin G2 (PGG2). The peroxidation stage is
the reduction stage of PGG2 into an unstable
endoperoxide compound called prostaglandin H2
(PGH2). There are two main isoforms of the
cyclooxygenase enzyme, cyclooxygenase-1 (COX-
1) and cyclooxygenase-2 (COX-2). COX-1 is
expressed continuously and has a function as a
regulator of homeostasis in the function of
protecting the gastric mucosa, maintaining platelet
integrity, and maintaining the function of renal
perfusion. COX-2 plays a role in pathologies such as
inflammation, pain, and cancer (Claria, 2003).
Molecular docking is a device that can be used to
study the interactions that occur from a molecular
complex. Molecular docking helps in studying drug
or ligand interactions with receptors or proteins.
Molecular docking is conducted by identifying the
corresponding active site of the receptor/protein,
obtaining the best geometry of the receptor ligand
and calculating the interaction energy of each
different ligand for designing a more effective
ligand. To perform molecular docking, the first thing
required is a three-dimensional structure of ligand
and receptor. Virtual screening is a computational
technique in the design of new computer-based
drugs (in silico) to identify the structures most likely
to bind to a targeted drug, usually a protein or
enzyme receptor (Mukesh and Rakesh, 2011).
2 MATERIALS AND METHODS
2.1 Materials
The tool used in this research was hardware and
software. Hard performances were equipped with
AMD E1-2100 APU with Radeon ™ HD Graphics
CPU GHz processor, 2GB RAM, and Microsoft
Windows 7 Ultimate 64-bit operating system, 24-
inch Hp® Monitor, and Bolt® modem for internet
access. The software programs were equipped with
the MGL Tools 1.5.6 Package consisting of
Autodock Vina, Autodock Tools, Pymol (DeLano
Scientific LLC.), Discovery Studio 4.5 Client, CLC
Drug Discovery Workbench 2.5, Chem office 2010,
Protein Data Bank (http://www.rcsb.org/pdb).
The material used was the 3D structure of the
platelet receptor that was downloaded from Protein
Data Bank which has formatted .pdb,i.e.
prostaglandin H2 synthase-1 (PDB ID: 1CQE) and
3D structure used was flavonoid derived compound
among others were eriocitrin, hesperidin,
neohesperidin, diosmin, rutin, luteolin, nobiletin,
sinensetin, and tangeritin (Molina et al., 2010).
2.2 Methods
Preparation of Prostaglandin H2 Synthase-1 (COX-
1) structure was conducted by downloading the
COX-1 receptor macromolecule from the Protein
Data Bank from http://www.rcsb.org/pdb formatted
from .pdb website to .pdb. Cavity must be
determined to find the residues in the receptor. The
cavity determination was performed using the
offline CLC Drug Discovery Workbench 2.5
software that was downloaded from
http://www.clcbio.com/products/clc-drug-discovery-
workbench/. Receptor macromolecules were
separated from solvents and ligands or non-standard
residues. The separation of macromolecules from
unnecessary molecules was done using the
Discovery Studio 4.0 program. The result of the
separation was saved in .pdb format. The design of
the ligand structure of the flavonoid derived
compound consists of eriocitrin, hesperidin,
neohesperidin, diosmin, rutin, luteolin, nobiletin,
sinensetin, and tangeritin were downloaded from the
PubChem site (http://pubchem.ncbi.nlm.nih.gov./).
The docking file preparation was conducted by
using Autodock Tools that was optimized by setting
the number of action torsion and converting the
format to .pdbqt. While the receptor preparation was
being conducted by adding hydrogen polar, the grid
MICH-PhD 2018 - 1st Muhammadiyah International Conference on Health and Pharmaceutical Development
20
box was set to know the position of the binding site
and the format was changed to .pdbqt. This file was
saved in a single folder in the C: drive on the
computer. Molecular Docking Process was
conducted using Autodock Vina. Ligands and
receptor that were already in drive C: copied and
converted in the form of notepad were saved with a
conf.txt name, Autodock Vina was executed with
command prompt program.
Molecular docking analysis was done by looking
at the free energy value of binding docking results,
viewed at the output in log.txt format. The selected
ligand-receptor complex was the complex which has
the lowest free binding energy value for further
analysis. The interaction between receptor and
ligand can be observed in Pymol software
3 RESULTS AND DISCUSSION
The macromolecule that was used as the docking
target was the Cyclooxygenase-1 enzyme (COX-1).
COX-1 was downloaded from the Protein Data
Bank. The PDB ID of COX-1 used was 1CQE
which has been used as a reference in predicting
antiplatelet activity based on previous research (Wu
et al., 2007). 1CQE consisted of 580 amino acid
residues. The 1CQE structure was downloaded from
the RCSB site with the format .pdb.
Cyclooxygenase-1 (COX-1) downloaded from the
RCSB must be cleaned using the offline Discovery
Studio 4.5 software because the receptor on RCSB is
holoprotein, which contains many ligands in the
receptor. The reason for the cleansing was to remove
the original disturbing ligands and water attached to
the receptor to speed up the docking process.
Molecular docking is carried out on the specific
region of the target protein, which is to be a binding
site. The location of this site is based on the ligand
or cofactor position co-crystallized with the structure
of the target protein, or the position of the amino
acids known for the binding position.
To get the cyclooxygenase-1 receptor inhibitory
effect, it must first recognize the residues that form
cavity and pocket in the target (receptor). The cavity
is a substance that is owned by the receptor. The
pocket is a space inside the cavity as access to the
bond between the ligand and the receptor, resulting
in the expected effect. Cavity search was carried out
using the CLC Drug Discovery Workbench 2.5
software located at the site
http://www.clcbio.com/products/clc-drug-discovery-
workbench/ (Glaab 2015). CLC Drug Discovery
Workbench 2.5 is a software managed by CLC Bio
A QIAGEN Company. This software can detect
bindings on receptors through the programs
provided.
Usually, more than one cavity is found in the
target receptor. Therefore, it was necessary to
evaluate the cavity to saw the possibility of the
cavity being a binding site that actually used the
CLC Drug Discovery Workbench 2.5 software. In
the search for COX-1 cavity receptors, many
cavities were detected in the COX-1 receptor regions
(Figure 1). It was necessary to do cavity evaluation
by setting a binding site on the Drug Discovery
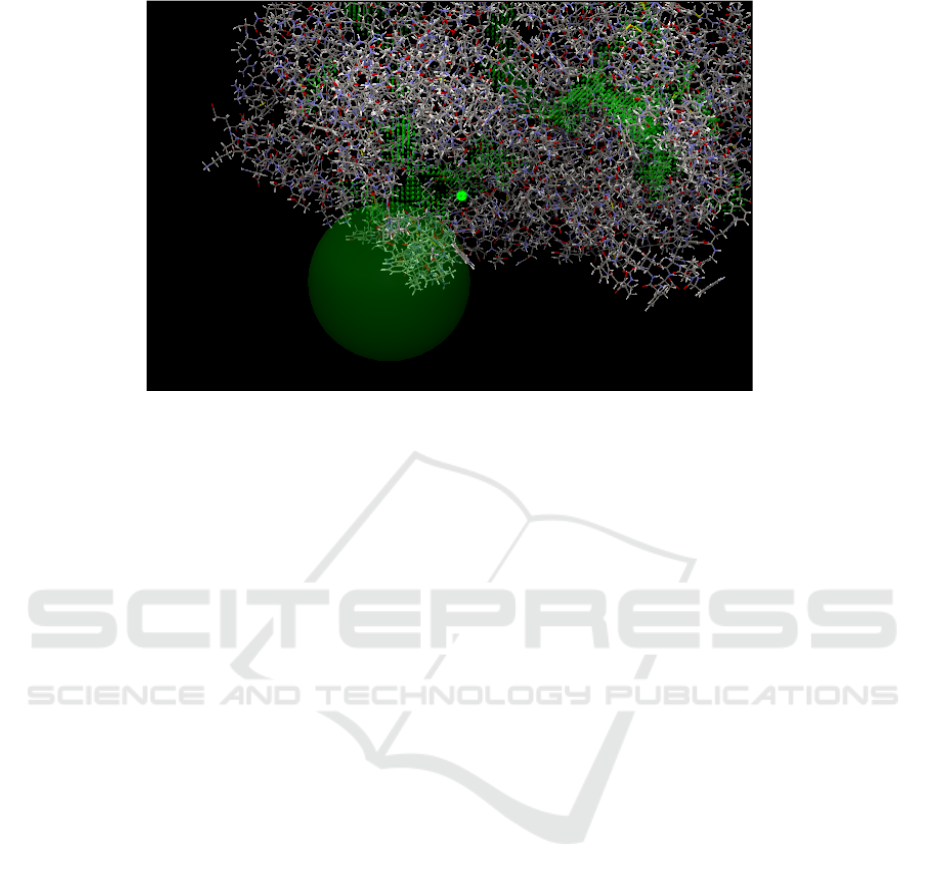
Figure1: Results of COX-1 receptor site binding detection (CLC Drug Discovery Software). Description: Areas marked
with green spots are cavity areas. The area in the white circle is the actual cavity as a binding site on the COX-1 receptor.
Molecular Docking Study of Lemon (Citrus limon (Linn) Burm. f) Flavonoid Derivatives Compound in Receptor Cyclooxygenase-1
(COX-1) as Antiplatelet in Ischaemic Stroke Disease
21
Workbench 2.5 CLC work program. After cavity
evaluation, the COX-1 receptor area that had been
detected by cavity showed a more specific area that
described the actual cavity as a binding site (Figure
1). This area is the place where interactions between
amino acid residues and receptors and ligands will
be used as the grid box area. The selection of cavity
binding pocket by CLC drug design based on the
largest pocket size (Li et al., 2008) and more open
and compact pockets have good properties for drug
binding (Cheng et al., 2007). Ligands, receptor, and
Autodock Vina software was saved in a folder
located on drive C: windows in the vina folder. The
destination was saved in one folder so that the
docking process can be carried out through the
command prompt. The command prompt is a
command line interface based program with written
work orders. At the receptor, the grid box must be
determined according to the results of the cavity
binding pocket from CLC Drug Design which was
set in the offline software of Autodock Tools. The
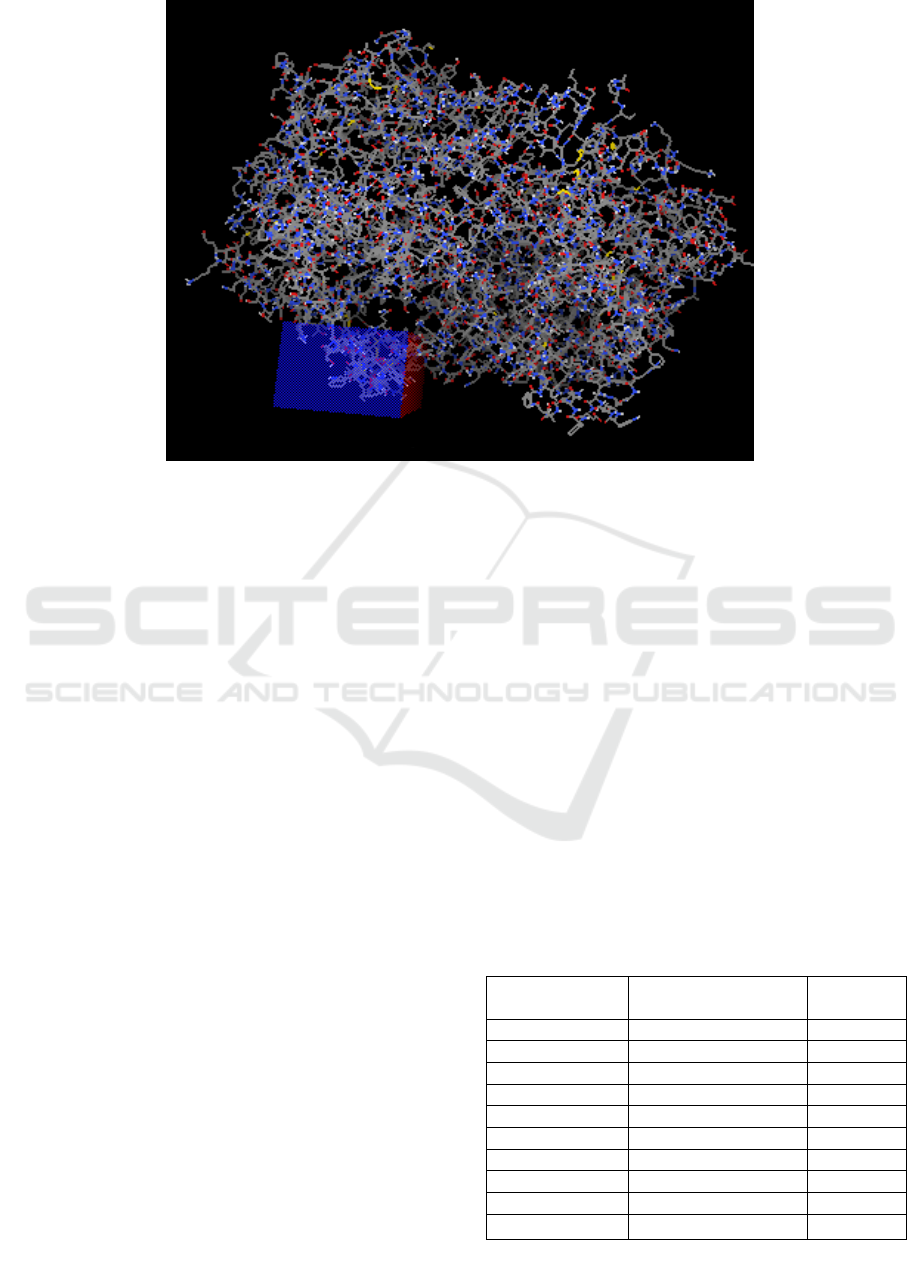
grid box used for 1CQE in the oriented docking
process was at the center_x coordinate = 28,453;
center_y = 7.271; center_z = 195.072, grid size 56 x
40 x 122Å with a spacing of 0.375Å (Figure 2).
Analysis of results in molecular docking includes
values of ΔG binding affinity and Root Mean Square
Deviation (RMSD). Molecular docking was
conducted to see the complex conformation of the
receptor-the docked ligand with Autodock Vina.
Determination of ligand conformation can be seen
from the results that come out in the command
prompt program which will be selected one of the
best out of nine conformations out of the docking
results by using Autodock Vina. The docking result
was the value of ΔG binding affinity (kcal/mol) for
one ligand. Affinity binding is a docking parameter
using Autodock Vina. The smaller the value of the
ΔG binding affinity, the affinity between the
receptor - the ligand will be higher and
otherwise, the
greater the value of ΔG binding affinity, the affinity
between receptor-ligand complex will be lower
(Rachmania et al., 2015).
Table 1: Results of standard ligand docking (Aspirin),
lemon plant ligand with COX-1 receptor (1CQE) using
Autodock Vina Software.
Ligands
ΔG Binding Affinity
(kcal/mol)
RMSD
(Å)
Aspirin -6.5 0
Neohesperidin -15.4 0
Rutin -15.3 0
Eriocitrin -14.9 0
Hes
p
eridin -14.7 0
Diosmin -14.1 0
Luteolin -10.0 0
Nobiletin -9.5 0
Sinensetin -9.5 0
Tangeritin -9.5 0
Figure 2: Results of COX-1 receptor site binding detection (CLC Drug Discovery Software And Autodock Tools).
Description: The area in the blue box is the actual cavity as a binding site on the COX-1 receptor.
MICH-PhD 2018 - 1st Muhammadiyah International Conference on Health and Pharmaceutical Development
22
Based on table 1, it can be seen that of the ten
ligands that were analyzed, the lowest values of ΔG
binding affinity in lemon are neohesperidin -15.4
kcal/mol and rutin -15.3 kcal/mol. The ΔG binding
value of affinity aspirin as a standard ligand is -6.5
kcal/mol. These values suggest that neohesperidin
and rutin ligands have better affinity than aspirin and
have antiplatelet potency. RMSD is the value used to
determine whether the prediction of the bond mode
is successful and important for validating the
docking program with a default value of ≤ 2Å. With
increasing deviations, the greater the error of
predicting the ligand interaction with receptors
(Brooijmans, 2009). The RMSD value obtained
from the docking of each ligand in the best
conformation is 0. This is caused by Vina compared
the value of each conformation with its best
conformation value.
The interaction between the receptor and the
ligand resulted in the distance between the bonds
Table 2: The distance of amino acid bond and residue, ligand functional group between Aspirin, Neohesperidin, and Rutin
with COX-1 Receptor (1CQE) using PyMol Software.
Ligands
The Distance of hydrogen
b
ond (Å)
Amino Acid Residue
Binding
Functional groups binding
Aspirin 2,8 Cys
41
-O
3,4 Cys
41
-O
Neohes
p
eridin 3,
0
Ar
g
469
-OH
3,1 Ar
g
469
-OH
3,5 Gln
44
-O
3,3 Cys
41
-OH
3,4 Cys
41
-OH
3,2 L
y
s
468
-OH
Rutin 3,4 L
y
s
468
-OH
2,3 Glu
465
-OH
3,0 Glu
465
-OH
2,8 Asp
135
-OH
3,2 Asp
135
-OH
3,2 Gl
y
45
-OH
3,4 Gl
y
45
-OH
2,6 C
y
s
41
-O
3,2 His
43
-OH
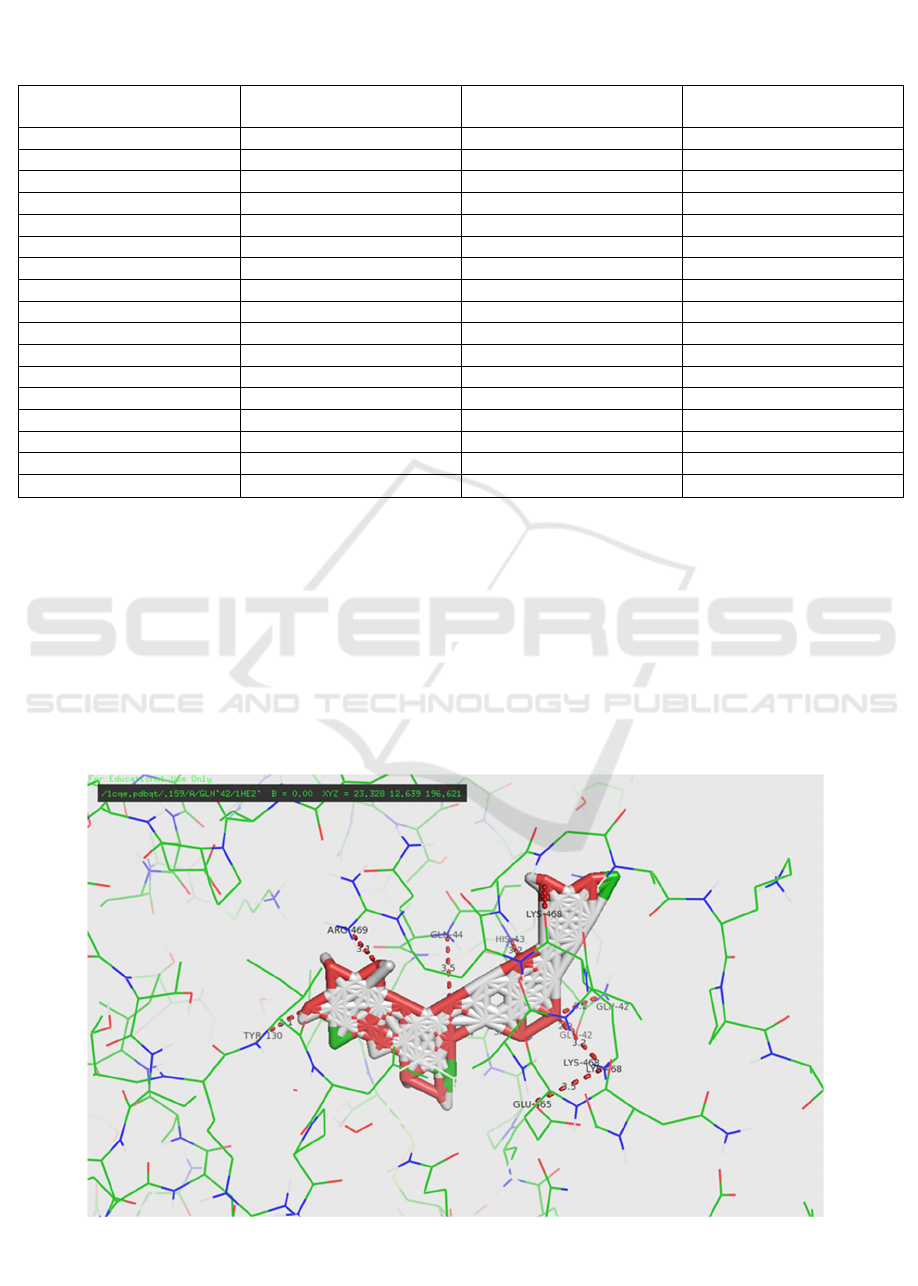
Figure 3. Residue contact between Neohesperidin and 1CQE receptor (Software Pymol)
Molecular Docking Study of Lemon (Citrus limon (Linn) Burm. f) Flavonoid Derivatives Compound in Receptor Cyclooxygenase-1
(COX-1) as Antiplatelet in Ischaemic Stroke Disease
23
and the bonded amino acid residues. Hydrogen
bonding is a bond that can occur by involving the
interaction of hydrogen atoms bonded covalently
with electronegative atoms such as Flour (F),
Nitrogen (N), Oxygen (O) (Glowacki et al., 2013).
Table 2 shows distances, amino acids and binding
groups that illustrate the interaction between
ligands/drugs with COX-1 receptors. The interaction
between aspirin as a standard drug against the COX-
1 receptor shows the presence of hydrogen bonds
from the -O group at Cys41 residues. For ligands
having the lowest ΔG binding affinity values,
neohesperidin and rutin, show that hydrogen bonds
are formed from the -O and -OH groups to the
Arg469, Gln44, Cys41, and Lys468 residues in
neohesperidin (Figure 3). The hydrogen bonds are
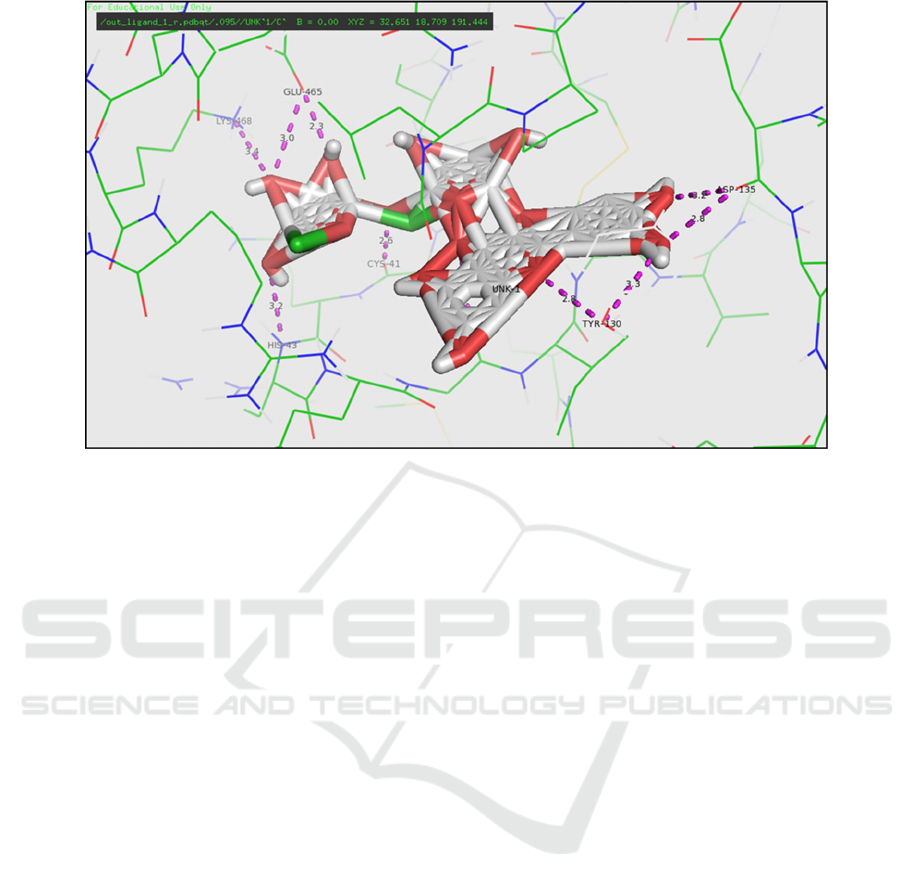
formed from the -O and -OH groups to the residues
Glu465, Cys41, Gly45, Asp135, Lys468, and His43
on the rutin (Figure 4). Hydrogen bonds may occur
between intermolecular and intramolecular, a good
range of hydrogen bonds at 2.5-3.5 Å (Syahputra et
al., 2014). The hydrogen bond distance that occurs
between amino acids residues in the COX-1 receptor
and neohesperidin and the rutin is a good hydrogen
bond because it is in the range 2.5-3.5 Å. So from
the results of the in silico analysis using molecular
docking method, it can be concluded that
neohesperidin and rutin compounds in lemon are
predicted to have potential as antiplatelet ischaemic
in stroke.
4 CONCLUSIONS
The flavonoid derived compounds found in lemon
(Citrus limon (Linn) Burm f) ie neohesperidin and
rutin result values of ΔG binding affinity (kcal/mol),
i.e. -15.4 kcal/mol and -15.3 kcal/mol. These
numbers are lower compared with aspirin
comparative drugs that has a value of ΔG binding
affinity -6.5 kcal/mol. neohesperidin and rutin
compounds have a better affinity than aspirin so that
it can be used as an antiplatelet candidate in
ischaemic stroke disease.
REFERENCES
Balitbang Kemenkes RI. 2013. Riset Kesehatan Dasar
(RISKESDAS). Balitbang Kemenkes RI. Jakarta.
Brooijmans N. 2009. Docking Methods, Ligand Design
and Validating Data Sets in The Structural Genomics
Era. Structural Bioinformatics Second Edition. Wiley
Interscience. New York. 645-646.
Claria J. 2003. Cyclooxygenase-2 Biology. Current
Pharmaceutical Design. 9 (27): 2177-2190.
Cheng, A. C., Coleman, R. G., Smyth, K. T., Cao, Q.,
Soulard, P., Caffrey, D. R., Salzberg, A. C., and
Huang, E. S. 2007. Structure-based maximal affinity
model predicts small-molecule druggability. Nat
Biotechnol, 25(1):71--75.
Chung CS, Caplan LR. 2007. Neurovascular Disorder.
Dalam: Christopher G, Goetz MD (eds). 2007.
Figure 4: Residual contact between Rutin ligand with 1cqe receptor (Software Pymol)
MICH-PhD 2018 - 1st Muhammadiyah International Conference on Health and Pharmaceutical Development
24

Textbook of Clinical Neurology 3rd edition. Saunders
Company. Philadelphia. 874 – 885.
Glaab E. 2015. Building A Virtual Ligand Screening
Pipeline Using Free Software. Briefing in
Bioinformatics. 17 (2): 352-356
Glowacki ED, Vladu MI, Bauer S. 2013. Hydrogen Bonds
in Molecular Solid From Biological Systems to
Organic Electronics. Journal of Material Chemistry.
(1): 3742-3753.
Gund, BM, Jagtap, PN, Ingale, VB, Patil, RY. 2013.
Stroke: A Brain Attack. IOSR Journal of Pharmacy.
3(8): 01-23.
Li, B., Turuvekere, S., Agrawal, M., La, D., Ramani, K.,
and Kihara, D. 2008. Characterization of local
geometry of protein surfaces with the visibility
criterion. Proteins, 71(2):670--683.
Middleton E, Kandaswami C, Theoharides TC. 2000.The
Effects of Plant Flavonoids on Mammalian Cells.
Pharmacological Reviews. 52(4): 673-751.
Molina EG, Perles RD, Moreno DA, Viguera CG. 2010.
Natural Bioactive Compounds of Citrus limon for
Food and Health. Journal of Pharmaceutical and
Biomedical Analysis. 51: 327-345.
Mukesh B, Rakesh K. 2011. Molecular Docking: A
Review. International Journal of Research in
Ayurveda and Pharmacy. 2(6): 1746-1751.
Rachmania, RA, Supandi, Larasati, OA. 2015. Analisis In-
Silico Senyawa Diterpenoid Lakton Herba Sambiloto
(Andrographis paniculata Nees) pada Reseptor Alpha-
Glucosidase Sebagai Antidiabetes Tipe II. Pharmacy.
12(02): 210-222.
Retnaningsih CH, Setiawan A, Sumardi. 2011. Potensi
Antiplatelet Kacang Koro (Mucuna pruriens L) dari
Fraksi Heksan dibandingkan dengan Aspirin pada
Tikus Hiperkolesterolemia. Seri Kajian Ilmiah. 14 (1):
80-86.
Riaz A, Khan RA, Mirza T. 2014. In vitro/In Vivo Effect
of (Citrus Limon L. Burm. f.) Juice on Blood
Parameters, Coagulation And Anticoagulation Factors
in Rabbits. Journal of Pharmaceutical Sciences. 27(4):
907-915.
Ringleb, PA, Bousser, MG, Ford, G, Bath, P, Brainin, M,
Caso, V, Cervera, Á, Chamorro, Al, Cordonnier, C,
Csiba, L. 2011. Ischaemic Stroke and Transient
Ischaemic Attack. European Handbook of
Neurological Management, Second Edition, Volume
1, Second Edition.
Syahputra, G, Ambarsari, L, Sumaryada, T. 2014.
Simulasi Docking Kurkumin Enol,
Bismetoksikurkumin dan Analognya Sebagai Inhibitor
Enzim12-Lipoksigenase. Jurnal Biofisika. 10(1): 55-
67.
Shinohara Y, Katayama Y, Uchiyama S, Yamaguchi T,
Handa S, Matsuoka K, Ohashi Y, Tanahashi N,
Yamamoto H, Genka C, Kitagawa Y, Kusuoka H,
Nishimaru K, Tsushima M, Koretsune Y, Sawada T,
Hamada C. 2010. Cilostazol for Prevention of
Secondary Stroke (CSPS 2): An Aspirin-Controlled,
Double-Blind, Randomised Non-Inferiority Trial. The
Lancet Neurology. 9(10): 959-968.
Wu CM, Wu SC, Chung WJ, Lin HC, Chen KT, Chen
YC, Hsu MF, Yang JM, Wang JP, Lin CN. 2007.
Antiplatelet Effect and Selective Binding to
Cyclooxygenase (COX) by Molecular Docking
Analysis of Flavonoids and Lignans. International
Journal of Molecular Sciences. 8: 830-841.
Molecular Docking Study of Lemon (Citrus limon (Linn) Burm. f) Flavonoid Derivatives Compound in Receptor Cyclooxygenase-1
(COX-1) as Antiplatelet in Ischaemic Stroke Disease
25
