
Vectorized Character Counting for Faster Pattern Matching
Roman Snytsar
Microsoft Research, One Microsoft Way, Redmond WA 98052, U.S.A.
Keywords:
Parallel Processing, Vectorization, Bioinformatics, FM-Index.
Abstract:
Many modern sequence alignment tools implement fast string matching using the space efficient data structure
called a FM-index. The succinct nature of this data structure presents unique challenges for the algorithm
designers. In this paper, we explore the opportunities for parallelization of the exact and inexact matches, and
present an efficient solution for the Occ portion of the algorithm that utilizes the instruction-level parallelism
of the modern CPUs. Our implementation computes all eight Occ values required for the inexact match
algorithm step in a single pass. We showcase the algorithm performance in a multi-core genome aligner and
discuss effects of the memory prefetch.
1 INTRODUCTION
The FM-index has been developed as a space efficient
index for string matching. The backward search over
the index finds exact matches of a pattern in time that
is linear relative to the length of the pattern, regard-
less the size of the reference. Even though the appli-
cations are numerous, for instance text compression
(Navarro and M
¨
akinen, 2007), and indexing (Zhang
et al., 2013a), the FM-index has become especially
popular with the developers of DNA sequence alig-
ners like Bowtie (Langmead et al., 2009), SOAPv2
(Li et al., 2009), and BWA (Li and Durbin, 2009).
Next, we introduce the fundamentals of the FM-index
construction and operation.
2 BACKGROUND
Let R be a string of length n over some alphabet Σ. A
special character $ that is not part of the alphabet and
is lexicographically smaller than any character in Σ is
appended to the end of the string. R[i] denotes a cha-
racter in R at position i, and R[i, j] is a substring of R
ranging from i to j. Suffix array SA(R) is then defined
as an integer array containing starting positions of all
suffixes of R in a sorted order so that
R[SA[i − 1], n] < R[SA[i], n], 1 < i ≤ n (1)
Suffix array could be constructed by simply sorting
all suffixes of a string. More sophisticated algorithms
take into account the fact that all strings are rela-
ted to each other and achieve much better asymptotic
complexity and practical performance (Puglisi et al.,
2007).
SA BWT Occ
A C G T
4 $ $ACA G 0 0 1 0
0 ACAG$ ACAG $ 0 0 1 0
2 AG$ AG$A C 0 1 1 0
1 CAG$ CAG$ A 1 1 1 0
3 G$ G$AC A 2 1 1 0
C= 0 2 3 4 4
Figure 1: Data structures comprising FM-index are in
boxes.
The Burrows-Wheeler Matrix BW M(R) is obtai-
ned by writing out all rotations of string R and sorting
them lexicographically. The last column of the BW M
then forms a string known as the Burrows-Wheeler
Transform BW T (R). Sorting string rotations is clo-
sely related to sorting prefixes as shown in Figure 1,
and BW T (R) can be easily obtained form SA(R):
BW T [i] =
(
R[SA[i] − 1], SA[i] > 0
$, SA[i] = 0
(2)
Next, for each character b in Σ and for every
0 ≤ k < n we record the number of occurrences of
b in the BW T substring BW T [0, k] , and store it in the
table Occ. Additionally, we store the total of occur-
rences of all characters lexicographically preceding b
Snytsar, R.
Vectorized Character Counting for Faster Pattern Matching.
DOI: 10.5220/0007258201490154
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 149-154
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
149

in BW T into the table C. It is easy to compute C as an
exclusive prefix sum of the last row of Occ. For any
single character at position i in a pattern W , the inter-
val of rows in the BWM starting with this character is
easily computed from C:
{k, l} = {C[W [i]],C[W [i] + 1]} (3)
From this initial interval, it is possible to extend the
search backward from the starting position using the
following recursive procedure:
E x a c tRe c u r (W, i , k , l )
i f i < 0 t h e n
r e t u r n
{
k, l
}
b ← W[i]
k ← C(b) + Occ(b, k − 1) + 1
l ← C(b) + Occ(b, l)
r e t u r n E xac t R e cur (W, i −1, k , l )
Listing 1: Backward Exact Match.
After the search is complete, the final BWT inter-
val is mapped back to the locations in reference using
the suffix array.
BWA (Li and Durbin, 2009) extends this algo-
rithm to allow a predetermined number of mismatches
z:
I n e xRe c u r (W, i , z , k , l )
i f z < 0 t h e n
r e t u r n ∅
i f i < 0 t h e n
r e t u r n
{
k, l
}
I ← ∅
I ← I
S
InexRecur(W, i − 1, z − 1, k, l)
f o r ea c h b ∈ Σ do
k ← C(b) + Occ(b, k − 1) + 1
l ← C(b) + Occ(b, l)
i f k ≤ l t h e n
I ← I
S
InexRecur(W, i, z − 1, k, l)
i f b = W[i] t h e n
I ← I
S
InexRecur(W, i − 1, z, k, l)
e l s e
I ← I
S
InexRecur(W, i − 1, z − 1, k, l)
r e t u r n I
Listing 2: Inexact Match.
Note that every step of the inexact match algo-
rithm consumes eight Occ values compared to two
Occ values required by the exact match. In theory,
all Occ values could be precomputed, but holding the
full Occ array for the human genome reference would
consume approximately 100GB of memory. To save
memory space, the FM-index over the DNA alphabet
is often stored using a cache-friendly approach intro-
duced in (Gog and Petri, 2014), that harkens back to
the bucket layout from the original FM-index paper
(Ferragina and Manzini, 2000). Values of Occ(∗, k)
for every k that is a multiple of 128 are stored in me-
mory followed by 128 characters of BWT in 2-bit en-
coding. Four Occ counters occupy 256 bits, as does
the BWT string. For Occ counters that are not at the
factor of 128 positions, the values must be calculated
on the fly. Furthermore, the suffix array is compres-
sed in a similar manner. Only values of SA[k] where k
is a multiple of 32 are stored in memory, while all the
values in between are recomputed using the Inverse
Suffix Array relationships:
Ψ
−1
(i) = C[BW T [i]] + Occ(BW T [i], i) (4)
SA[k] = SA[(Ψ
−1
)
j
(k)] + j (5)
It means that Equation 4 is applied over and over until
for a value of j the result comes out to be a multiple of
32, and the SA value could be constructed according
to Equation 5.
Even though the memory saving measures do not
change the asymptotic complexity of the match algo-
rithm, in reality they add hundreds of computations of
Occ to every search. Given that the search is perfor-
med multiple times for each and every read out of bil-
lions required for the alignment of a human genome,
Occ function performance becomes crucial.
3 SOLUTION
Our approach could be traced back to the algorithm
by (Vigna, 2008) that performs memory table look-
ups to count character occurrences in each byte of the
BWT string. We replace the memory lookups with
the half-byte register lookups, building on an idea first
proposed by Mula for the bit population count (Muła
et al., 2017). Note however that we do not attempt
to reduce the character counting problem to bit coun-
ting, and apply the half-byte technique directly to the
BWT string. Inputs and outputs for all eight counters,
that are required for one step of the inexact search, fit
into a single AVX512 register and are computed with
one vector pass.
The input BWT string is masked with zeros befo-
rehand for situations when the character at position
k is in the middle of the byte. The result of 256-
bit occurrence count should be corrected for the extra
127 − k A characters.
3.1 Lookup
Every byte in the BWT string is split into its higher
and lower half. Since each half byte value cannot
be greater than 15, the lookup values now fit into a
single vector register and could be retrieved via the
VPSHUFB instruction. The lookup returns all four
counters in a 2-bit format packed into a byte. Two bits
are sufficient as a half byte contains just two charac-
ters. Additionally, the OccLo result is pre-converted
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
150
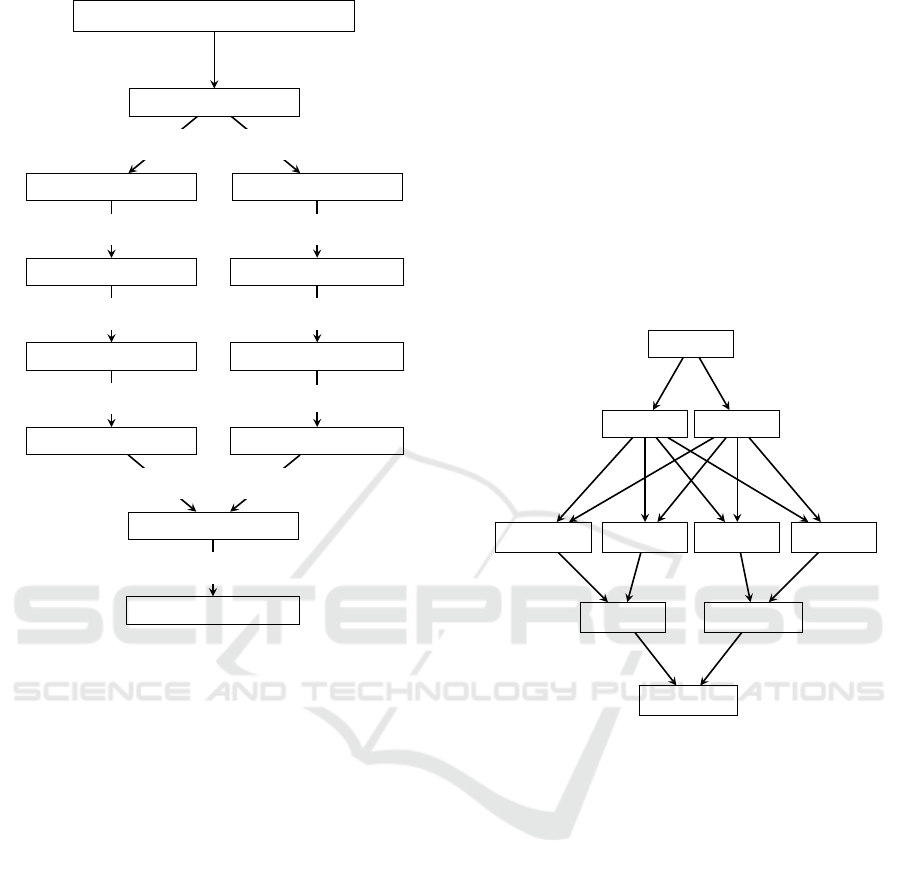
”ccacttgcgaaatttacaaggtttattaggtt”
fa3cfe813f026f45
0a0c0e010f020f05 0f030f0803000604
dfbeaffa7fee7ff7 8041801141021405
0dfbeaffa7fee7ff
0804180114102140
fdfffefffffeffff
0000000100000100
00000000000007f2
0000000000000006
vpand vpsrld, vpand
vpshufb vpshufb
vpsrlvq
vpsrlvq
vpandnot
vpor
vpsadbw
vpsadbw
vpsubq
Figure 2: Example of counting letter ’g’ in a 32-character
portion of BWT string.
into its bit complement to assist with subsequent ex-
traction operation.
3.2 Extraction
After the lookup phase, all counters are tightly pac-
ked into two vector registers. Any addition operation
could result in overflowing the 2-bit values. Before
proceeding we have to extract counters for a given
character X.
Both OccLo and OccHi are shifted to bring Occ
X
into the two lower bits of the byte. High bits of OccLo
are then filled with ones and high bits of OccHi
with zeros. At this point OccHi contains unsigned
byte values of OccHi
X
, and OccLo contains values
of 255 − OccLo
X
. The merged values are then fed
into the VPSADBW instruction. It sums the absolute
differences of eight consecutive bytes and stores the
result as a 64-bit integer. At the end of this operation
the result vector contains four partial sums in the form
of 2040−Σ(OccLo
X
+ OccHi
X
). The final horizontal
sum yields 8160 − Occ
X
, and the final subtraction is
combinable with the correction for the extra A count.
3.3 Aggregation
The extraction sequence runs four times to collect par-
tial sums for characters ACGT, CATG, TGCA, and
GTAC in four vector registers. Aggregating four final
sums within a single register then takes just three ad-
ditions and three shuffle operations, only one of which
crosses the 128-bit lane boundary. The absence of
data dependencies between extraction operations fa-
cilitates efficient use of the SIMD pipeline and keeps
all arithmetic ports busy. Outputs for all eight coun-
ters required for one step of the inexact search fit into
a single AVX512 register and are computed with one
vector pass.
BWT
Lo
Hi
G,T,A,CT, G, C, A C,A,T,G A,C,G,T
G,T,A,C A, C, G, T
A, C, G, T
Lookup
Extract
Aggregate
Figure 3: Data shuffling.
4 IMPLEMENTATION
We have implemented the half-byte Occ algorithm
using the AVX2 instruction set. The AVX512 ver-
sion computing 8 values in parallel has also been inte-
grated in the inexact search algorithm. The assembly
code along with the Intel Architecture Code Analy-
zer throughput report is listed in the Appendix. The
code is indeed well balanced: 49 out of 65 micro ope-
rations are evenly spread across the ports 0, 1, and 5
available for the arithmetics and shuffles. The code is
expected to bottleneck on the backend meaning that
the memory access pattern is crucial for the real word
performance. High utilization of every bit and the
branchless nature of the algorithm make it also a good
candidate for implementing on a field-programmable
gate array hardware (FPGA).
Vectorized Character Counting for Faster Pattern Matching
151
4.1 Experimental Setup
The computer platform is an Intel Xeon Platinum
8168 system with 16 cores running at 2.7 GHz and
32GB of RAM. To test the software performance we
have run the BWA alignment tool with 16 threads
(-t 16) on a 30X Human genome sample NA12878
from the 1000 Genomes database using hg38 as a re-
ference. We have executed the BWA version 7.15 to
establish the baseline, and then replaced the Occ code
with our AVX2 and AVX512 implementations. The
total run times got collected from the BWA reports,
and the percentage of time spent in the BWT and SA
code versus the rest of BWA have been measured via
profiling.
4.2 Results
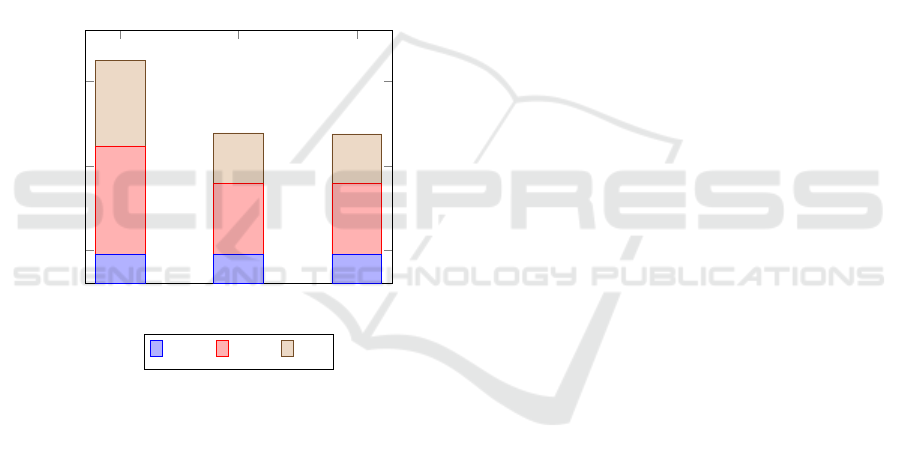
Scalar AVX2 AVX512
1
2
3
·10
4
Execution time, s
Other BWT SA
Figure 4: Execution times for NA12878.
The vectorized SA code runs twice as fast as the
scalar version, mostly due to the predictable memory
prefetch pattern. The BWT code exhibits only a 30%
speedup, and the switch to AVX512 does not bring
any gains. At this point the BWT code is comple-
tely memory bound. To utilize the vectorization per-
formance gains completely, we would have to ex-
plore ideas for improving the cache locality of the
FM-index (Chacon et al., 2013; Zhang et al., 2013b).
Despite the memory bottlenecks, the overall runtime
is improved by 25%.
REFERENCES
Chacon, A., Moure, J. C., Espinosa, A., and Hernandez, P.
(2013). n-step fm-index for faster pattern matching.
Procedia Computer Science, 18:70 – 79. 2013 Inter-
national Conference on Computational Science.
Ferragina, P. and Manzini, G. (2000). Opportunistic data
structures with applications. In 41st Annual Sym-
posium on Foundations of Computer Science, pages
390–398. IEEE.
Gog, S. and Petri, M. (2014). Optimized succinct data struc-
tures for massive data. Software: Practice and Expe-
rience, 44(11):1287–1314.
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L.
(2009). Ultrafast and memory-efficient alignment of
short dna sequences to the human genome. Genome
biology, 10(3):R25.
Li, H. and Durbin, R. (2009). Fast and accurate short read
alignment with burrows–wheeler transform. Bioinfor-
matics, 25(14):1754–1760.
Li, R., Yu, C., Li, Y., Lam, T.-W., Yiu, S.-M., Kristian-
sen, K., and Wang, J. (2009). Soap2: an improved
ultrafast tool for short read alignment. Bioinformatics,
25(15):1966–1967.
Muła, W., Kurz, N., and Lemire, D. (2017). Faster popu-
lation counts using avx2 instructions. The Computer
Journal, 61(1):111–120.
Navarro, G. and M
¨
akinen, V. (2007). Compressed full-text
indexes. ACM Comput. Surv., 39(1).
Puglisi, S. J., Smyth, W. F., and Turpin, A. H. (2007).
A taxonomy of suffix array construction algorithms.
ACM Comput. Surv., 39(2).
Vigna, S. (2008). Broadword implementation of rank/select
queries. In International Workshop on Experimental
and Efficient Algorithms, pages 154–168. Springer.
Zhang, D., Liu, Q., Wu, Y., Li, Y., and Xiao, L. (2013a).
Compression and indexing based on bwt: A survey.
In 2013 10th Web Information System and Application
Conference, pages 61–64.
Zhang, J., Lin, H., Balaji, P., and Feng, W.-c. (2013b). Op-
timizing burrows-wheeler transform-based sequence
alignment on multicore architectures. In Cluster,
Cloud and Grid Computing (CCGrid), 2013 13th
IEEE/ACM International Symposium on, pages 377–
384. IEEE.
APPENDIX
Intel Architecture Code Analyzer report for the AVX
implementation of Ψ
−1
.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
152

Intel(R) Architecture Code Analyzer Version - v3.0-28-g1ba2cbb build date: 2017-10-23;17:30:24
Analyzed File - bwa.exe
Binary Format - 64Bit
Architecture - SKL
Analysis Type - Throughput
Throughput Analysis Report
--------------------------
Block Throughput: 20.63 Cycles Throughput Bottleneck: Backend
Loop Count: 22
Port Binding In Cycles Per Iteration:
--------------------------------------------------------------------------------------------------
| Port | 0 - DV | 1 | 2 - D | 3 - D | 4 | 5 | 6 | 7 |
--------------------------------------------------------------------------------------------------
| Cycles | 16.3 0.0 | 16.4 | 7.5 7.5 | 7.5 7.5 | 0.0 | 16.3 | 1.0 | 0.0 |
--------------------------------------------------------------------------------------------------
|Num Of| Ports pressure in cycles | |
| Uops| 0 | 1 | 2 - D | 3 - D | 4 | 5 | 6 | 7 |
---------------------------------------------------------------
| 2ˆ | | 1.0 | 0.5 0.5 | 0.5 0.5 | | | | | vpaddq xmm1, xmm10, xmmword ptr [rip+0x21dff0]
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | mov r8, qword ptr [rdx]
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu xmm2, xmmword ptr [rip+0x21dfd5]
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm6, ymmword ptr [rip+0x21dfad]
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm7, ymmword ptr [rip+0x21e105]
| 1 | | | | | | 1.0 | | | vmovq xmm0, r8
| 1 | | | | | | 1.0 | | | vpbroadcastq xmm0, xmm0
| 1 | | | | | | 1.0 | | | vpcmpgtq xmm0, xmm0, xmm10
| 2 | 1.0 | 1.0 | | | | | | | vpblendvb xmm3, xmm10, xmm1, xmm0
| 1 | 0.3 | 0.7 | | | | | | | vpandn xmm0, xmm2, xmm3
| 1 | 0.7 | 0.3 | | | | | | | vpsrlq xmm1, xmm0, 0x1
| 1 | 0.3 | 0.7 | | | | | | | vpand xmm9, xmm3, xmm2
| 1 | 1.0 | | | | | | | | vmovq rcx, xmm1
| 2ˆ | | | 0.5 0.5 | 0.5 0.5 | | | 1.0 | | add rcx, qword ptr [rdx+0x40]
| 1 | | | | | | 1.0 | | | vpbroadcastb ymm4, xmm9
| 1 | | 1.0 | | | | | | | vpandn ymm0, ymm4, ymm6
| 1 | 0.7 | 0.3 | | | | | | | vpsrld ymm1, ymm0, 0x17
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm5, ymmword ptr [rcx+0x20]
| 1 | 0.3 | 0.7 | | | | | | | vpsrlvd ymm2, ymm5, ymm1
| 1 | 0.7 | 0.3 | | | | | | | vpsrld ymm3, ymm4, 0x1c
| 1 | 0.3 | 0.7 | | | | | | | vpsrad ymm0, ymm7, 0x18
| 1 | | | | | | 1.0 | | | vpandn ymm1, ymm0, ymm2
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu xmm0, xmmword ptr [rip+0x21e01c]
| 1 | | | | | | 1.0 | | | vpermd ymm2, ymm3, ymm1
| 1 | 0.7 | 0.3 | | | | | | | vpslld ymm8, ymm2, 0x1
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm2, ymmword ptr [rip+0x21e01a]
| 1 | 0.3 | 0.7 | | | | | | | vpslld xmm1, xmm4, 0x1
| 1 | 0.7 | 0.3 | | | | | | | vpsubusb xmm1, xmm0, xmm1
| 1 | | | | | | 1.0 | | | vpmovsxbd ymm3, xmm1
| 1 | 0.3 | 0.7 | | | | | | | vpsllvd ymm0, ymm2, ymm3
| 1 | 0.4 | 0.3 | | | | 0.3 | | | vpand ymm4, ymm0, ymm5
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm0, ymmword ptr [rip+0x21df5b]
| 1 | 0.3 | 0.4 | | | | 0.3 | | | vpand ymm1, ymm4, ymm6
| 1 | | | | | | 1.0 | | | vpshufb ymm1, ymm0, ymm1
| 1 | 0.7 | 0.3 | | | | | | | vpsrlvd ymm2, ymm1, ymm8
| 1 | | | 0.5 0.5 | 0.5 0.5 | | | | | vmovdqu ymm1, ymmword ptr [rip+0x21df85]
| 1 | 0.3 | 0.7 | | | | | | | vpor ymm5, ymm2, ymm7
| 1 | 0.7 | 0.3 | | | | | | | vpsrld ymm0, ymm4, 0x4
| 1 | | 0.3 | | | | 0.7 | | | vpand ymm3, ymm0, ymm6
| 1 | | | | | | 1.0 | | | vpshufb ymm2, ymm1, ymm3
| 1 | 0.6 | 0.4 | | | | | | | vpsrlvd ymm0, ymm2, ymm8
| 1 | 0.4 | 0.6 | | | | | | | vpandn ymm3, ymm7, ymm0
Vectorized Character Counting for Faster Pattern Matching
153

| 1 | | | | | | 1.0 | | | vpsadbw ymm1, ymm3, ymm5
| 2ˆ | 0.6 | 0.4 | 0.5 0.5 | 0.5 0.5 | | | | | vpaddd ymm5, ymm8, ymmword ptr [rip+0x21dffe]
| 1 | | | | | | 1.0 | | | vpermilpd ymm0, ymm1, 0x5
| 1 | 0.4 | 0.6 | | | | | | | vpaddq ymm4, ymm0, ymm1
| 2ˆ | 0.6 | 0.4 | 0.5 0.5 | 0.5 0.5 | | | | | vpaddw xmm0, xmm9, xmmword ptr [rip+0x21debc]
| 1 | | | | | | 1.0 | | | vpmovsxwq ymm1, xmm0
| 2ˆ | 0.4 | 0.6 | 0.5 0.5 | 0.5 0.5 | | | | | vpaddq ymm2, ymm1, ymmword ptr [rdx+0x8]
| 2ˆ | 0.6 | 0.4 | 0.5 0.5 | 0.5 0.5 | | | | | vpaddq ymm3, ymm2, ymmword ptr [rcx]
| 1 | | | | | | 1.0 | | | vpermq ymm0, ymm4, 0x4e
| 1 | 0.4 | 0.6 | | | | | | | vpaddq ymm1, ymm0, ymm4
| 1 | | | | | | 1.0 | | | vmovq xmm0, r8
| 1 | 0.6 | 0.4 | | | | | | | vpsubq ymm2, ymm3, ymm1
| 1 | | | | | | 1.0 | | | vpermd ymm4, ymm5, ymm2
| 1 | 0.4 | 0.6 | | | | | | | vpcmpeqq xmm1, xmm10, xmm0
| 1 | 0.6 | 0.4 | | | | | | | vpandn xmm0, xmm1, xmm4
| 1 | 1.0 | | | | | | | | vmovq r8, xmm0
Total Num Of Uops: 65
Analysis Notes:
Backend allocation was stalled due to unavailable allocation resources.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
154
