
Plant Diseases Recognition from Digital Images using Multichannel
Convolutional Neural Networks
Andre da Silva Abade
1
, Ana Paula G. S. de Almeida
2
and Flavio de Barros Vidal
3
1
Federal Institute of Education, Science and Technology of Mato Grosso, Brazil
2
Department of Mechanical Engineering, University of Brasilia, Distrito Federal, Brazil
3
Department of Computer Science, University of Brasilia, Distrito Federal, Brazil
Keywords:
Convolutional Neural Networks, Multichannel Convolutional Neural Networks, Plant Disease, Crop Disease
Recognition, Computer Vision.
Abstract:
Plant diseases are considered one of the main factors influencing food production and to minimize losses in
production, it is essential that crop diseases have a fast detection and recognition. Nowadays, recent studies use
deep learning techniques to diagnose plant diseases in an attempt to solve the main problem: a fast, low-cost
and efficient methodology to diagnose plant diseases. In this work, we propose the use of classical convoluti-
onal neural network (CNN) models trained from scratch and a Multichannel CNN (M-CNN) approach to train
and evaluate the PlantVillage dataset, containing several plant diseases and more than 54,000 images (divided
into 38 diseases classes with 14 plant species). In both proposed approaches, our results achieved better accu-
racies than the state-of-the-art, with faster convergence and without the use of transfer learning techniques.
Our multichannel approach also demonstrates that the three versions of the dataset (colored, grayscaled and
segmented) can contribute to improve accuracy, adding relevant information to the proposed artificial neural
network.
1 INTRODUCTION
Plant diseases are considered one of the main factors
influencing food production, being responsible for the
significant reduction of the physical or economic pro-
ductivity of the crops and, in certain cases, may be
an impediment to this activity. According to Altieri
(2018), in order to minimize production losses and
maintain crop sustainability, it is essential that dise-
ase management and control measures be carried out
in an appropriate manner, highlighting the constant
monitoring of the crop, combined with the rapid and
accurate diagnosis of the diseases. These practices are
the most recommended by phytopathologists.
The major challenge for agriculture is the correct
identification of the symptoms of major diseases that
affect crops (Anderson et al., 2004). Manual and me-
chanized practices in traditional planting processes
are not able to cover large areas of plantation and pro-
vide essential early information to decision-making
processes (Miller et al., 2009). Thus, it is necessary
to develop automated solutions, practical, reliable and
economical able to monitor the health of plants provi-
ding meaningful information to the decision-making
process (e.g. correct dosage of pesticides (Mahlein,
2016)).
Computer Vision along with Artificial Intelligence
(AI) has been developing techniques and methods
for recognizing and classifying objects with signifi-
cant advances (Arnal Barbedo, 2013). These systems
use Convolutional Neural Networks (CNNs) (Lecun
et al., 1998) and their results in some experiments
are already superior to humans in large-scale recon-
naissance tasks. The studies presented in Mohanty
et al. (2016) and Ferentinos (2018) make use of deep
learning techniques in agriculture, in particular in
the diagnosis of plant diseases. These approaches
have used two popular architectures, namely Alex-
Net (Krizhevsky et al., 2012) and GoogLeNet (Incep-
tion v1) (Szegedy et al., 2014), which were designed
in the context of the Scale Visual Recognition Chal-
lenge (ILSVRC) (Russakovsky et al., 2014) for the
ImageNet dataset (Deng et al., 2009).
With the aforementioned architectures, Mohanty
et al. (2016) show that only the colored dataset is suf-
ficient to recognize plant diseases. However, more in-
formation about a subject can contribute to improve
the network accuracy and to confirm this assump-
450
Abade, A., S. de Almeida, A. and Vidal, F.
Plant Diseases Recognition from Digital Images using Multichannel Convolutional Neural Networks.
DOI: 10.5220/0007383904500458
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 450-458
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

tion, other versions – gray-scale and segmented – of
the PlantVillage (Hughes et al., 2015) dataset were
combined into multichannel convolutional neural net-
works, using the same architectures to our models for
a fair comparison. Furthermore, our work also im-
proves the reference single channel baseline without
using transfer learning techniques.
The paper is organized as follows: In Section 2
are presented the related works about plant disease
recognition. Section 3 explains our methodology.
Section 4 contains information about the results and
discussion. Section 5 provides conclusion points and
further works.
2 RELATED WORDS
The recognition and classification of leaf diseases of
plants is a problem with many challenges to over-
come. The analysis in the process of identification
of the diseases through the leaves can incur a large
number of false positives, for example, the symptoms
of phytotoxicity are associated with some disease due
to similar leaf lesions.
We developed an extensive time-review (from
most to less recent) of the main literature works, from
the traditional techniques and methods used in the
process of recognition and classification of foliar di-
seases in plants to the latest advances provided by the
use of Convolutional Neural Networks (CNNs), sin-
gle and multichannel approaches. Table 1 presents
these works in chronological order, summarizing the
used techniques and methods and their consequent
area of application.
Before the advent of CNNs, traditional machine
learning classification methods, such as SVM (Rumpf
et al., 2010) and K-Means (Al-hiary et al., 2011),
were used to classify diseases in plants. Patil
and Bodhe (2011) applied classic image processing
technique for disease detection in sugarcane leaves by
using threshold segmentation to determine leaf area
and triangle threshold for lesioning area, getting the
average accuracy of 98.60%. An approach proposed
by Singh and Misra (2017) uses genetic algorithms
for image segmentation which is an important aspect
for disease detection in a plant leaf.
Relevant works approach feature extraction
techniques for the detection of plant diseases. It is
possible to highlight the studies of Pydipati et al.
(2006), where there is use of color co-occurrence
method (CCM) to determine whether texture based
hue, saturation, and intensity (HSI) color features in
conjunction with statistical classification algorithms
could be used to identify diseased and normal citrus
leaves under laboratory conditions. The leaf sam-
ple discriminant analysis using CCM textural featu-
res achieved classification accuracies of over 95% for
all classes when using hue and saturation texture fe-
atures. According to Jabal et al. (2013), feature ex-
traction is a promising approach capable of solving
dichotomies between datasets constructed with ima-
ges in controlled environments and images captured
in the real world. This study proposed an ideal case
approach in plant classification and recognition that
was not only applicable in the real world, but also
acceptable in laboratory conditions.
Due to the increase in processing capacity trig-
gered by the use of Graphics Process Unit (GPU),
AI is corroborating significantly with the robust set
of traditional resources applied by Computer Vision
techniques (Ferentinos, 2018). Tacitly, Machine Le-
arning techniques have demonstrated significant gains
in accuracy in the process of classification and identi-
fication of plant diseases.
These advances are demonstrated in the works
of Rumpf et al. (2010), which proposes an approach
for the detection and differentiation of plant disea-
ses using Support Vector Machine algorithms. In this
study, the authors implemented a technique to iden-
tify beet diseases, in which depending on the type
and stage of disease the classification accuracy was
between 65% and 90%. Another approach based on
leaf images and using Artificial Neural Networks as
a technique for an automatic detection and classifica-
tion of plant diseases was used in conjunction with
K-means as a clustering procedure proposed in the
works of Al-hiary et al. (2011). On average, the accu-
racy of classification using this approach was 94.67%.
According to LeCun et al. (2015), deep learning
allows computational models to learn representations
of data with multiple levels of abstraction, improving
the state-of-the-art in many domains, such as speech
recognition, object recognition, object detection. One
particular type of deep, feedforward network that was
much easier to train and generalized much better than
networks with full connectivity was the convolutio-
nal neural networks (CNNs). CNNs constitute one
of the most powerful techniques for modeling com-
plex processes and performing pattern recognition in
applications with large amount of data, like the one
of pattern recognition in images (LeCun et al., 2015).
Sladojevic et al. (2016) develops a model using CNN
capable of recognizing 13 different types of diseases
of healthy leafy plants, with the ability to distinguish
the leaves of the plants from their surroundings. The
experimental results on the developed model achieved
precision between 91% and 98%, for separate class
tests, on average 96.3%.
Plant Diseases Recognition from Digital Images using Multichannel Convolutional Neural Networks
451

Table 1: Review on the methods and techniques of leaf plant diseases’ recognition and classification.
Year Author Method Application area
2018 Ferrentinos Convolutional neural network Identification of leaf disease from 25 different species of plants
2018 Lin Zhongqi et al. Multichannel Convolutional neural network Detecting maize leaf diseases for 5 diseases
2017 Yang Lu et al. Convolutional neural network Identification of rice diseases
2017 Pawara et al. Local descriptors and CNN Identification of fruits diseases
2017 Tallha Akram et al. Based on an Image processing technique Real time classification of plant diseases
2017 Trimi Neha Tete et al. Neural network, K-means and thresholding Identification of disease from potato, apple and mango leaves
2017 Vijai Singh et al. Image segmentation technique Detection of plant leaf diseases
2017 Megha S. et al. Fuzzy c means and Support vector machine Identification of plant leaf disease
2017 Lin Yuan et al. Fisher’s linear discriminant analysis (FLDA) Identification of plant diseases and pests form SAR images
2016 Mohanty et al. Convolutional neural network Identification of leaf disease from 25 different species of plants
2016 Sladojevic et al. Convolutional neural network Identification of plant leaf disease
2016 Pujari et al.
Support vector machine and
Artificial neural network
Identification of plant leaf disease of crops such as wheat,
maize, grape, sunflower etc.
2016 Ramakrishnan M. et al. Backpropagation algorithm Identification of groundnut leaf disease
2016 Malvika Ranjan et al. Artificial neural network Identification of cotton leaf disease
2015 Praksh M. Mainkar et al.
K-means clustering, GLCM and
Backpropagation neural network
Identification of disease from potato, tomato and cotton leaves
2014 Marion Neumann et al. Support vector machine Identification of beet leaf disease
2014 Rong Zhou et al. Support vector machine Identification of Cercospora Leaf Spot from Sugar beet
2013 Jabal et al. Features extraction Recognition and classification of plant leaf disease
2011 Patil et al. Based on an Image processing technique Identification of plant leaf disease
2011 Al-hiary et al. K-menas clustering Identification of plant leaf disease
2010 T. Rumpf et al. Support vector machine Identification of Sugar beet disease from leaves
2006 Pydipati et al. Color texture features Identification of Citrus disease
Using a public dataset of 54,306 images of disea-
sed and healthy plant leaves collected under control-
led conditions, the study of Mohanty et al. (2016)
train a deep neural network to identify 14 crop spe-
cies and 26 diseases (or absence thereof). The trained
model achieves an accuracy of 99.35% on a held-out
test set, demonstrating the feasibility of this appro-
ach. Pawara et al. (2017) compared the performance
of some conventional pattern recognition techniques
with that of CNN models, in plants identification,
using three different databases of images of either en-
tire plants and fruits, or plant leaves, concluding that
CNNs drastically outperform conventional methods.
The propose described in Lu et al. (2017) pre-
sents a novel rice diseases identification method ba-
sed on CNN techniques. Using a dataset of 500 natu-
ral images of diseased and healthy rice leaves, CNNs
are trained to identify 10 common rice diseases. Un-
der the 10-fold cross-validation strategy, the proposed
CNNs-based model achieves an accuracy of 95.48%.
Finally, Ferentinos (2018) used CNN models with an
open database of 87,848 images, containing 25 dif-
ferent plants in a set of 58 distinct classes of [plant,
disease] combinations, including healthy plants. Se-
veral model architectures were trained, with the best
performance reaching a 99.53% success rate in identi-
fying the corresponding [plant, disease] combination
(or healthy plant).
In the context of a M-CNN approach, Lin et al.
(2018) describes a simple use of the M-CNN archi-
tecture to detect and recognize maize leaf diseases,
using a dataset of 10,820 images containing five com-
mon maize leaf diseases. This approach uses a Re-
gion of Interest (ROI) to preprocess the input image
and achieves an accuracy of 92.31% with 30,000 ite-
rations/epochs. Even though this result was not better
than all of the single channel CNNs approaches des-
cribed earlier in this Section, the use of a reduced da-
taset in Lin et al. (2018) indicates that a M-CNN may
be a relevant and improved approach for plant disea-
ses detection and recognition.
3 PROPOSED METHODOLOGY
The research process of this study was guided by the
work of Mohanty et al. (2016). The state-of-the-art
shown by the author motivated our efforts to improve
not only the accuracy achieved by the methods pre-
viously proposed, but also develop and implement an
approach to produce more consistent results. In Mo-
hanty et al. (2016) work, it is shown that the colored
dataset is sufficient to perform the recognition of plant
diseases. Our work combines in a multichannel con-
volutional neural network (M-CNN) the other availa-
ble versions of the dataset in the same model in or-
der to improve the network accuracy. Also, our work
improves the single channel CNN’s baseline without
using transfer learning techniques. The chosen trai-
ning/testing ratio was 80/20, the ratio that produced
better results in the reference work.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
452

3.1 Dataset
The proposed methodology uses the PlantVillage da-
taset, provided by Hughes et al. (2015), containing
54,306 images of plant leaves and 38 different classes,
each class corresponding to a different crop disease.
Every class has three different versions: original co-
lored image, grayscaled image and segmented image.
Figures 1-(A) to (C) show a sample of this dataset.
Figure 1: Examples of plant diseases Dataset: (A) The three
versions of apple scab disease. (B) A sample of black rot, a
grape disease. (C) Three versions of strawberry leaf scorch.
3.2 CNN Architecture
According to Goodfellow et al. (2016), CNNs are spe-
cialized artificial neural networks that process input
data with some kind of spatial topology, such as ima-
ges, videos, audio and text. In addition to convo-
lution layers, CNNs are usually composed of other
types of layers, such as pooling. In this work, two
classic CNNs are used to evaluate the dataset: Alex-
Net (Krizhevsky et al., 2012) and GoogLeNet (Incep-
tion v1) (Szegedy et al., 2014).
3.2.1 Multichannel CNNs
Multichannel CNNs (M-CNNs) are generally used
when parallel processing of the input data is desi-
red (Karpathy et al., 2014). Such streams can eventu-
ally merge into one in the latter layers of the network.
In the relevant contributions of the studies of Baccou-
che et al. (2011) and Ji et al. (2013), it is common
for the point of concatenation to be present before the
first fully connected layer of the network, that is, the
parallel processing is concentrated between the con-
volution layers. In Karpathy et al. (2014), a 2-channel
CNN is proposed, each channel receiving two frames
of the input video and being capable of generating la-
bels of the main action. Another advantage of using
M-CNNs is also highlighted by Karpathy et al. (2014)
and it consists in reducing the dimensionality of the
network input, which helps to decrease the proces-
sing time. In Figure 2 it is presented a generic archi-
tecture of a network with two input channels. Each
channel receives one different type of the dataset, ge-
nerating three additional versions:Version 1: Color +
Grayscale; Version 2: Color + Segmented; Version
3: Grayscale + Segmented.
There were not any pre-processing steps and all
the images had the same resolution size of 256 × 256
pixels. The objective behind the use of multichannel
networks is to observe whether the neural network can
produce better results if additional information is pro-
vided. Our models use a late fusion technique (Kar-
pathy et al., 2014), where two separate single channel
networks with shared parameters are merged in the
first fully connected layer, computing global features
by comparing outputs of both streams.
To improve the single channel baseline, a hyperpa-
rameter optimization strategy was used. The optimi-
zation of the hyperparameters in the training of CNNs
is a process that demands a lot of effort, due to the nu-
merous parameters that can be adjusted, to the context
of the input data, to the deep learning network model
used and the defined architecture. In this study we
adopted the grid search capability to adjust the hyper-
parameters of each learning model. Our hyperpara-
meter optimization strategy makes use of a reference
value as the starting point for exploring a range of va-
lues according to each parameter that can be adjusted.
The grid search was constructed with reference to the
values presented in study of Mohanty et al. (2016).
Thus, specific values were selected to achieve the best
results during training.
According to Srivastava et al. (2014), dropout
is a regularization technique for reducing overfit-
ting in neural networks by preventing complex co-
adaptations on training data.
Basically, half of the neurons on a particular layer
will be deactivated during training. The generaliza-
tion is improved due to the forcing of your layer to
learn the same ”concept” with different neurons. Nor-
mally, some deep learning models use dropout on the
fully connected layers, but is also possible to use dro-
pout after the max-pooling layers, creating some kind
of image noise augmentation.
In our approach, dropout layers are added before
and after the fusion that occurs in M-CNN architec-
Plant Diseases Recognition from Digital Images using Multichannel Convolutional Neural Networks
453

Figure 2: An illustration of the generic architecture of a multichannel convolutional network. The model generalizes a
structure with two input channels and identifies the most relevant segments of architecture.
ture. We insert a dropout layer between the pooling
layers that precede the fusion of the networks and af-
ter the fusion, for the first two fully connected layers.
In addition, based on the observations about the va-
lues of loss, we adjust the fraction of inputs to 0 at
each update during training time, which helps prevent
overfitting.
All CNN models (single and multichannel) were
trained using the training parameters presented in Ta-
ble 2.
Transfer learning is the technique of training
a base network on a base dataset, usually Image-
Net (Deng et al., 2009), and then transfer the learned
features to a second target network to be trained on
a target dataset and task (Yosinski et al., 2014). Fre-
quently, this approach tends to improve network over-
all accuracy, as seen in the results of Mohanty et al.
(2016). One of our goals in this work was to out-
perform the state-of-the-art values without the use of
transfer learning.
According to Pan and Yang (2010), transfer lear-
ning techniques are advantageous when used in CNNs
because they shorten training time since initial weig-
hts are imported from a similar training experience
performed on a larger data set. Thus, it is possible
to increase the accuracy of a CNN using transfer le-
arning even though its dataset is noticeably smaller.
However, it should be noted that if the dataset fea-
tures to be used has unique peculiarities to its set of
objects and the input images have different dimensi-
ons of pre-trained model, the use of learning transfer
technique should be rethought.
In the development of this study, after analyzing
the data set with its relevant characteristics, the mo-
del and architecture of each convolutional neural net-
work used, we chose starting the learning process
from scratch.
Table 2: Training hyperparameters.
Model Hyperparameters Values
AlexNet
Learning rate 0.01
Momentum 0.9
Weight decay 1e-6
Batch size 128
GoogleNet
Learning rate 0.0205
Momentum 0.9
Weight decay 0.0005
Batch size 16
4 RESULTS
This section presents the results obtained based on
the CNN architectures detailed in the methodology.
The GPU used for training the proposed models was a
NVIDIA GeForce GTX Titan Xp and all models were
developed using TensorFlow API version 1.6 (Abadi
et al., 2015) and Keras version 2.2.1 (Chollet et al.,
2015) frameworks. For evaluation, we used mean F
1
score and overall accuracy.
For better visualization, AlexNet and GoogleNet
with multichannel architecture will be named as M-
AlexNet and M-GoogleNet.
Table 3 shows the results of the proposed metho-
dology for single and multichannel architectures after
computing the mean F
1
score of each network, with
the best achieved result highlighted. In addition, we
present the best results obtained in the work of Mo-
hanty et al. (2016) as a reference for discussions. All
multichannel models were trained from scratch.
Figures 3 to 10 show the performance and losses
of all the testing models during the training process.
In Section 3, we report that we would use the trai-
ning process from scratch to compute the results. In
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
454

Table 3: Mean F
1
score of the proposed architectures and a comparison.
Model Dataset Type Mean F
1
Score
Mohanty et al. (2016)
AlexNet (transfer learning) Color 0.9927
GoogleNet (transfer learning) Color 0.9934
Ours
AlexNet (from scratch) Color 0.9873
GoogleNet (from scratch) Color 0.9940
M-AlexNet (from scratch)
Version 1 0.9959
Version 2 0.9920
Version 3 0.9923
M-GoogleNet (from scratch)
Version 1 0.9955
Version 2 0.9938
Version 3 0.9941
Table 3, our results are presented with significant im-
provements, even though we do not use transfer lear-
ning techniques.
Basically, it is possible to explain this impro-
vement of results for the single channel from scratch
networks in comparison to the same proposal of the
work of Mohanty et al. (2016), by the singularity of
the characteristics of the dataset. Although the dataset
is considered small by its number of images, the mo-
dels pre-trained with the imaging do not have a signi-
ficant sample space of labeled diseased plants. There-
fore, although the training time increased subtly, the
accuracy gains were representative.
In the study of Mohanty et al. (2016) its con-
clusions make it understood that the grayscaled and
segmented versions do not collaborate for an impro-
vement of the accuracy when comparing to the co-
lored version of the dataset. In our evaluation after
the experiments, it was possible to observe that for
models with single channel architecture that assump-
tion remains consistent, even though they were trai-
ned from scratch and with hyperparameters optimiza-
tion.
However, when we use M-CNN networks by mer-
ging the 3 different versions of the dataset into a 2-
channel architecture, we again explore the unique ex-
traction of characteristics from each version of the da-
taset, improving the learning of the model. Our appro-
ach has demonstrated that each version of the dataset
enriches the learning of the model, promoting a signi-
ficant gain in accuracy.
Our results using the approach with multichan-
nel networks tacitly demonstrated that networks with
simpler architectures, such as AlexNet, obtained hig-
her accuracy to a network with denser architecture.
With only hyperparameter adjusting, our best single
channel result is better than 0.06% of the best result
obtained by Mohanty et al. (2016).
Considering the use of additional versions of the
dataset, grayscaled and segmented images, our best
result is better than 0.25% of the state-of-the-art va-
lue and it even outperforms our single channel met-
hod. The best combination output was with Version
1 of the dataset, that is colored and grayscaled images.
The overall results of M-CNNs were consistent with
any of the two different models, AlexNet and Goog-
leNet.
The graphics in Figures 3 up to 10 show that the
networks appear to stabilize after 30 epochs, but the
application of more epochs could increase the achie-
ved results.
Figure 3: Single channel networks accuracy on testing da-
taset.
Figure 4: Single channel networks losses on testing dataset.
5 CONCLUSIONS
In this work, we explored the potentialities of the
convolutional neural networks already evidenced by
the literature to identify plant diseases through sam-
ples from healthy and diseased plants. We explored
Plant Diseases Recognition from Digital Images using Multichannel Convolutional Neural Networks
455
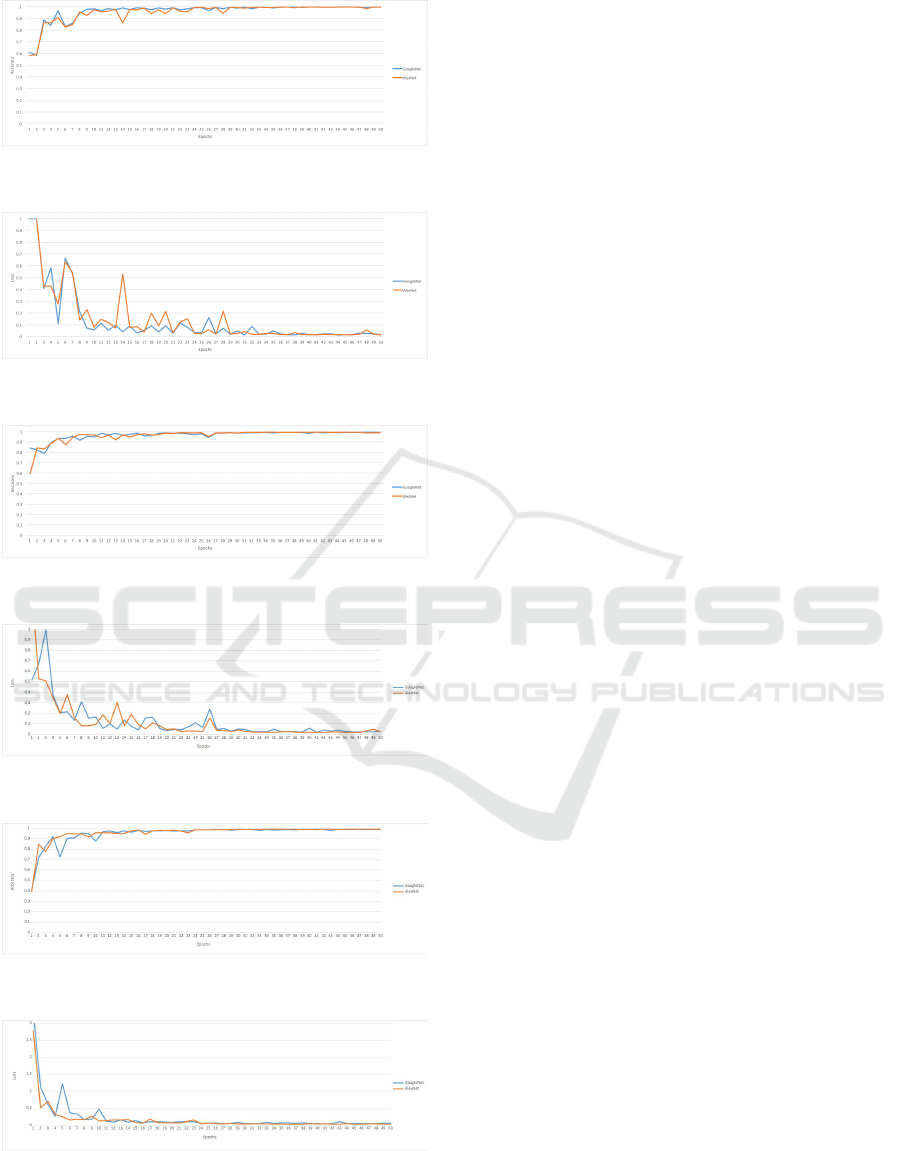
Figure 5: Multichannel networks accuracy on testing data-
set for Version 1.
Figure 6: Multichannel networks losses on testing dataset
for Version 1.
Figure 7: Multichannel networks accuracy on testing data-
set for Version 2.
Figure 8: Multichannel networks losses on testing dataset
for Version 2.
Figure 9: Multichannel networks accuracy on testing data-
set for Version 3.
Figure 10: Multichannel networks losses on testing dataset
for Version 3.
primarily knowledge gaps highlighted by Mohanty
et al. (2016) optimizing and improving their results
and proposing an approach using convolutional neu-
ral networks with multichannels. The training of the
models was performed using an openly available da-
tabase PlantVillage, consisting of 54,306 images con-
taining 38 classes. We adopted the same strategy
as Mohanty et al. (2016) when performing the training
with three preprocessed versions of the PlantVillage
dataset defined as color, grayscale and segmented.
In the first step of our approach, we achieved sig-
nificant advances as the accuracy of single channel
networks, optimizing the hyperparameters and adjus-
ting the dropout layers according to the dataset cha-
racteristics to minimize overfitting. It should be noted
that knowing the gains of transfer learning techniques,
we chose to train from scratch in order to demonstrate
the possibility of customization and gains in the lear-
ning process compared to a sample considered small
for a dataset.
Also, the additional inputs of the network provide
an even better accuracy, showing that M-CNNs were
able to enhance the general system, generating the
best overall result in this work and keeping the mean
F
1
scores regular and robust, independently of the
chosen model. The reference model achieved 0.9934
while our M-CNN obtained 0.9959. The dataset avai-
lable did not have images of plants in cultivated envi-
ronments, so the results of our approach only contem-
plate the tests performed extremely under preproces-
sed images and acquired in controlled environments.
Overall, we can conclude that a M-CNN model
trained from scratch is better than a single channel
model with transfer learning in two aspects: faster
convergence and reduced processing time. Further-
more, other image frequencies (e.g. grayscale) are
crucial to improve the general accuracy. Also, when
we train a single convolutional neural network from
scratch we achieve a model 10 times smaller than a
single channel with transfer learning. This reduction
allow us to build applications using real time plant di-
sease identification in an open field using mobile de-
vices.
In future works, the next step is to apply this ap-
proach to a dataset of images of healthy and disea-
sed plants obtained in growing environments. Thus,
it will be possible to adjust the M-CNN approach to
meet the requirements for identifying and classifying
plant diseases in culture from new real collected ima-
ges.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
456

ACKNOWLEDGEMENTS
The authors also express their gratitude and ackno-
wledge the support of NVIDIA Corporation with the
donation of the Titan Xp GPU used for this research
and the financial support received from the Federal In-
stitute of Education, Science and Technology of Mato
Grosso.
REFERENCES
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z.,
Citro, C., Corrado, G. S., Davis, A., Dean, J., De-
vin, M., Ghemawat, S., Goodfellow, I., Harp, A., Ir-
ving, G., Isard, M., Jia, Y., Jozefowicz, R., Kaiser,
L., Kudlur, M., Levenberg, J., Man
´
e, D., Monga, R.,
Moore, S., Murray, D., Olah, C., Schuster, M., Shlens,
J., Steiner, B., Sutskever, I., Talwar, K., Tucker, P.,
Vanhoucke, V., Vasudevan, V., Vi
´
egas, F., Vinyals, O.,
Warden, P., Wattenberg, M., Wicke, M., Yu, Y., and
Zheng, X. (2015). TensorFlow: Large-scale machine
learning on heterogeneous systems. Software availa-
ble from tensorflow.org.
Al-hiary, H., Bani-ahmad, S., Reyalat, M., Braik, M., and
Alrahamneh, Z. (2011). Fast and accurate detection
and classification of plant diseases. International
Journal of Computer Applications, 17(1):31 – 38.
Altieri, M. (2018). Agroecology: The Science Of Sustai-
nable Agriculture. CRC Press, Endereo, 2nd edition
edition.
Anderson, P. K., Cunningham, A. A., Patel, N. G., Morales,
F. J., Epstein, P. R., and Daszak, P. (2004). Emerging
infectious diseases of plants: pathogen pollution, cli-
mate change and agrotechnology drivers. Trends in
Ecology & Evolution, 19(10):535 – 544.
Arnal Barbedo, J. G. (2013). Digital image processing
techniques for detecting, quantifying and classifying
plant diseases. SpringerPlus, 2(1):660.
Baccouche, M., Mamalet, F., Wolf, C., Garcia, C., and Bas-
kurt, A. (2011). Sequential deep learning for human
action recognition. In International Workshop on Hu-
man Behavior Understanding, pages 29–39. Springer.
Chollet, F. et al. (2015). Keras.
https://github.com/fchollet/keras.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). ImageNet: A Large-Scale Hierarchical
Image Database. In CVPR09.
Ferentinos, K. P. (2018). Deep learning models for plant
disease detection and diagnosis. Computers and Elec-
tronics in Agriculture, 145:311 – 318.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. MIT Press. http://www.deeplearningbook.
org.
Hughes, D., Salath
´
e, M., et al. (2015). An open access re-
pository of images on plant health to enable the deve-
lopment of mobile disease diagnostics. arXiv preprint
arXiv:1511.08060.
Jabal, M. F. A., Hamid, S., Shuib, S., Ahmad, I., Jabal, M.
F. A., Hamid, S., Shuib, S., and Ahmad, I. (2013).
Leaf Features Extraction and Recognition Approa-
ches To Classify Plant. Journal of Computer Science,
9(10):1295–1304.
Ji, S., Xu, W., Yang, M., and Yu, K. (2013). 3d convolu-
tional neural networks for human action recognition.
IEEE transactions on pattern analysis and machine
intelligence, 35(1):221–231.
Karpathy, A., Toderici, G., Shetty, S., Leung, T., Sukthan-
kar, R., and Fei-Fei, L. (2014). Large-scale video
classification with convolutional neural networks. In
CVPR.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012).
Imagenet classification with deep convolutional neu-
ral networks. In Pereira, F., Burges, C. J. C., Bottou,
L., and Weinberger, K. Q., editors, Advances in Neu-
ral Information Processing Systems 25, pages 1097–
1105. Curran Associates, Inc.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep lear-
ning. Nature, 521(7553):436–444.
Lecun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998).
Gradient-based learning applied to document recogni-
tion. Proceedings of the IEEE, 86(11):2278–2324.
Lin, Z., Mu, S., Shi, A., Pang, C., and Sun, X. (2018). A
novel method of maize leaf disease image identifica-
tion based on a multichannel convolutional neural net-
work. Transactions of the ASABE, 0(0):0.
Lu, Y., Yi, S., Zeng, N., Liu, Y., and Zhang, Y. (2017).
Identification of rice diseases using deep convolutio-
nal neural networks. Neurocomputing, 267:378 – 384.
Mahlein, A.-K. (2016). Plant disease detection by ima-
ging sensors–parallels and specific demands for preci-
sion agriculture and plant phenotyping. Plant Disease,
100(2):241–251.
Miller, S. A., Beed, F. D., and Harmon, C. L. (2009). Plant
Disease Diagnostic Capabilities and Networks. An-
nual Review of Phytopathology, 47(1):15–38.
Mohanty, S., Hughes, D., and Salath, M. (2016). Using
deep learning for image-based plant disease detection.
Frontiers in Plant Science, 7(September).
Pan, S. J. and Yang, Q. (2010). A survey on transfer le-
arning. IEEE Transactions on Knowledge and Data
Engineering, 22(10):1345–1359.
Patil, S. B. and Bodhe, S. K. (2011). Leaf disease severity
measurement using image processing.
Pawara, P., Okafor, E., Surinta, O., Schomaker, L., and Wie-
ring, M. (2017). Comparing local descriptors and bags
of visual words to deep convolutional neural networks
for plant recognition. In Proceedings of the 6th Inter-
national Conference on Pattern Recognition Applica-
tions and Methods, ICPRAM 2017, Porto, Portugal,
February 24-26, 2017., pages 479–486.
Pydipati, R., Burks, T., and Lee, W. (2006). Identification of
citrus disease using color texture features and discri-
minant analysis. Computers and Electronics in Agri-
culture, 52(1):49 – 59.
Rumpf, T., Mahlein, A.-K., Steiner, U., Oerke, E.-C.,
Dehne, H.-W., and Plmer, L. (2010). Early detection
and classification of plant diseases with support vector
Plant Diseases Recognition from Digital Images using Multichannel Convolutional Neural Networks
457

machines based on hyperspectral reflectance. Compu-
ters and Electronics in Agriculture, 74(1):91 – 99.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S.,
Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bern-
stein, M. S., Berg, A. C., and Li, F. (2014). Image-
net large scale visual recognition challenge. CoRR,
abs/1409.0575.
Singh, V. and Misra, A. (2017). Detection of plant leaf di-
seases using image segmentation and soft computing
techniques. Information Processing in Agriculture,
4(1):41 – 49.
Sladojevic, S., Arsenovic, M., Anderla, A., Culibrk, D., and
Stefanovic, D. (2016). Deep neural networks based
recognition of plant diseases by leaf image classifica-
tion. Computational Intelligence and Neuroscience,
2016.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I.,
and Salakhutdinov, R. (2014). Dropout: A simple way
to prevent neural networks from overfitting. Journal
of Machine Learning Research, 15:1929–1958.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., An-
guelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A.,
et al. (2014). Going deeper with convolutions.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? In Ghahramani, Z., Welling, M., Cortes, C.,
Lawrence, N. D., and Weinberger, K. Q., editors, Ad-
vances in Neural Information Processing Systems 27,
pages 3320–3328. Curran Associates, Inc.
VISAPP 2019 - 14th International Conference on Computer Vision Theory and Applications
458
