
Mean and Variability in RNA Polymerase Numbers Are Correlated
to the Mean but Not the Variability in Size and Composition of
Escherichia Coli Cells
Bilena Almeida, Vatsala Chauhan, Vinodh Kandavalli and Andre Ribeiro
1
Laboratory of Biosystem Dynamics, BioMediTech Institute and Faculty of Biomedical Sciences and Engineering,
Tampere University of Technology, Tampere, Finland
Keywords: RNA Polymerase, Cell-to-cell Variability, Flow Cytometry, Single-cell Biology, Statistical Analysis.
Abstract: Cell morphology differs with cell physiology in general and with gene expression in particular. We
investigate the degree to which these relationships differ with medium richness. Using Escherichia coli cells
with fluorescently tagged β’ subunits, flow cytometry, and statistical analysis, we study at the single-cell
level the correlation between parameters associated to cell morphology and composition (FSC, SSC, and
Width channels) and GFP tagged RNA polymerase (RNAp) levels (FITC channel). From measurements in
three media differing in richness (M63, LB, and TB) and, thus, cell growth rates, we find that the mean and
cell-to-cell variability in RNAp levels are correlated to the mean values of FSC, SSC, and/or Width.
Further, in all growth conditions considered, RNAp levels are positively correlated to FSC, SSC, and Width
at the single-cell level, with the correlation decreasing for increasing medium richness. Overall, the results
suggest that the mean and cell-to-cell variability in levels of RNAp, a master regulator of gene expression,
are correlated to the mean values of the parameters assessing the cellular morphology and composition, as
measured by flow cytometry, but they do not correlate to the degree of variability of these parameter values.
1 INTRODUCTION
In Escherichia coli, the concentration of RNA
polymerases (RNAp) is a key regulator of the rate of
transcription (McClure, 1980, 1985; Arkin, Ross and
McAdams, 1998; Kærn et al., 2005; Browning and
Busby, 2016). As this concentration differs even
between sisters cells (Cabrera and Jin, 2003;
Bratton, Mooney and Weisshaar, 2011; Yang et al.,
2014), it is an extrinsic factor for cell-to-cell
variability in gene expression (Elowitz et al., 2002;
Mäkelä, Kandavalli and Ribeiro, 2017).
One source of cell-to-cell variability in RNAp
numbers is the noise in the chemical processes
responsible for the production of RNAp (see e.g.
(Gillespie, 1977)). Other sources include variability
in cells’ health, morphology, and components
(Elowitz et al. 2002; Muthukrishnan et al., 2014;
Oliveira et al., 2016).
Here, we investigate the degree to which the
morphology and composition of the cells of a
population correlate with their mean and variability
in RNAp numbers. Since the environment is known
to affect the morphology and composition, we study
how this correlation differs with medium richness.
For this, we use E. coli strain RL1314 which has
GFP tagged β’ subunits (Bratton, Mooney and
Weisshaar, 2011). To assess both fluorescence levels
as well as parameters associated to cells’
morphology and composition, we use Flow
cytometry. Measurements are conducted in M63,
LB, and TB media, where growth rates differ. From
the measurements, we collect data on the cells’
green fluorescence intensity levels (a proxy for
RNAp numbers), and on the cells’ morphology
(size) and composition. Using the data, we searched
for statistically significant correlations between the
RNAp levels and morphology and composition, in
media differing in richness.
226
Almeida, B., Chauhan, V., Kandavalli, V. and Ribeiro, A.
Mean and Variability in RNA Polymerase Numbers Are Correlated to the Mean but Not the Variability in Size and Composition of Escherichia Coli Cells.
DOI: 10.5220/0007456102260233
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 226-233
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

2 METHODS
2.1 Bacterial Cells, Chemicals, Growth
Conditions, and Growth Rates
We used E. coli RL1314 with fluorescently (GFP)
tagged β’ subunits (Bratton, Mooney and Weisshaar,
2011), generously provided by Robert Landick,
University of Wisconsin-Madison, U.S.A.. For cell
cultures, chemicals components for Luria-Bertani
(LB), terrific broth (TB) and M63 media were
purchased from LabM (UK) and Sigma-Aldrich.
Casamino acids and vitamins were purchased from
Gibco. LB medium components are 1 g tryptone, 0.5
g yeast extract and 1 g NaCl (pH – 7.0). Meanwhile,
the composition of TB medium per 100ml is 1.2 g
tryptone, 2.4 g yeast extract, 0.4% glycerol and TB
salts (KH2PO4 and K2HPO4). M63 medium was
prepared using M63 salts supplemented with 0.4%
glycerol, vitamins and 20% casamino acids.
Prior to flow cytometry, RL1314 cells were
grown overnight at 30 ºC with aeration and shaking
in the appropriate medium, diluted 1:1000 into the
fresh specific medium and allowed to grow at 37 ºC
at 250 rpm until an optical density at 600 nm
(OD
600
) of 0.4. Growth rates were measured by
growth curves obtained from cells at 37°C in the
appropriate medium (LB, TB and M63) with
antibiotics, using a spectrophotometer (Ultrospec 10;
GE Health Care). Cultures were grown overnight at
30°C with aeration and shaking at 250 rpm. Next,
overnight cultures were diluted into fresh medium to
an initial OD
600
of 0.01. The OD
600
values were
monitored every 20 min for 3.2 h.
2.2 Flow Cytometry
For flow cytometry (FC), cells from 5 ml of
bacterial culture were diluted 1:10000 into 1 ml PBS
vortexed for 10 seconds and a total of 50.000 cells
were tested in each run. Prior to every day
experiments, the analyzer was calibrated using
ACEA NovoCyte particle QC beads
(Cat.No.8000004). Data was collected using an
ACEA NovoCyte Flow Cytometer (ACEA
Biosciences Inc., San Diego USA) equipped with a
blue laser (488 nm) for excitation and the
fluorescein isothiocyanate channel (FITC) (530/30
nm filter) for detecting emitted light at a flow rate of
14 µl/minute and a core diameter of 7.7 µM. A PMT
voltage of 417 was used for FITC. To avoid
background signal from particles smaller than
bacteria, the detection threshold was set to 5000 in
FSCH analyses.
From the flow cytometry data, we study: i) FITC,
which measures the green fluorescence intensity
from a cell (a proxy for the number of RNA
polymerases in the cell); ii) Forward scatter (FSC),
which measures the light scattered at less than 10
degrees as a cell passes through the laser beam (a
proxy for cell size); iii) Side scatter (SSC), which
measures the light scattered at a 90 degree angle as a
cell passes through the laser beam (a proxy for cell
density); and, iv) Width (W), which measures the
duration of the signal, not impacted by the PMT
voltage, which also correlates with cell size. Except
for the Width, the FC informs on both the ‘Height’
(H) and ‘Area’ (A) of the signals. The H is the
maximum peak of the signal while the A is the
integration of the H measures over time.
Note that, in all conditions, we removed from the
data any cell with a negative or abnormally high or
low parameter value (which amounted to ~10-15%
of the cells in each medium condition). This is
necessary since, when ignoring one of the
parameters, the correlation between this and the
remaining ones cannot be obtained.
2.3 Correlations
Correlations between parameters extracted by FC
are obtained by linear regressions using the least-
squares fit method (95% confidence intervals),
applying the Matlab function fitlm that creates a
LinearModel object. We obtain the coefficient of
determination (R
2
) of the fitted regression line for
each case, along with the P-value of statistical
significance (derived from the F-test under the null
hypothesis that all regression coefficients equal
zero). If this P-value is smaller than 0.01, we reject
the null hypothesis that the line is a constant i.e., that
one variable does not differ with the other.
3 RESULTS
We investigate whether the cells’ morphology and
composition parameters measured by FC (FSC,
SSC, and Width channels) are correlated with RNAp
levels (FITC channel), and whether these
correlations differ with medium richness.
3.1 Growth Rates
We placed cells in LB (control), M63, and TB
media. For differences between conditions to be
significant,
cells should differ significantly in mean
Mean and Variability in RNA Polymerase Numbers Are Correlated to the Mean but Not the Variability in Size and Composition of
Escherichia Coli Cells
227
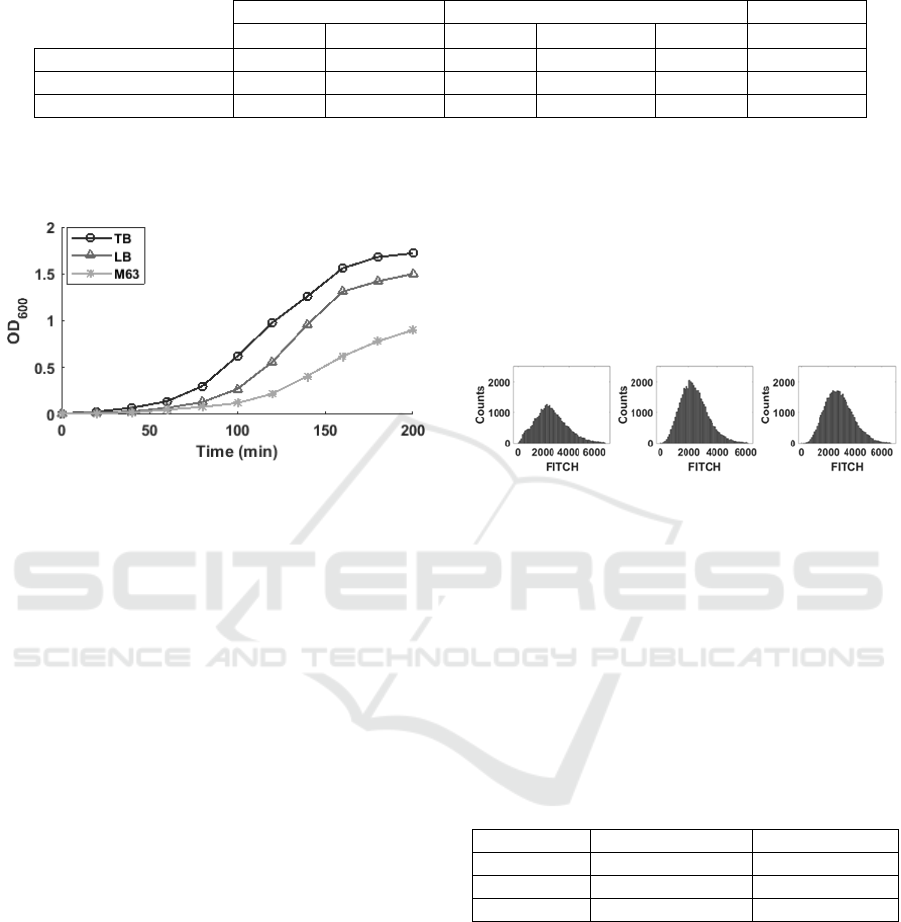
Table 1: Correlation (R
2
) between Height (H) and Area (A) for FITC, FSC and SSC in each medium.
M63 LB TB
R
2
P-value R
2
P-value R
2
P-value
FITCA vs FITCH 0.86 <0.01 0.78 <0.01 0.73 <0.01
FSCA vs FSCH 0.94 <0.01 0.84 <0.01 0.83 <0.01
SSCA vs SSCH 0.98 <0.01 0.94 <0.01 0.96 <0.01
growth rates. This differences were verified in this
OD
600
measurements.
Figure 1: Growth curves of cells of the RL1314 strain in
various media, as measured by OD
600
.
From Figure 1, M63, the poorest medium, has
the slowest growth rate, followed by LB and, finally,
TB, the richest medium with the fastest growth rate,
as expected from previous studies (see e.g.
(Goncalves et al, 2018)).
3.2 Correlation between Height (H)
and Area (A) of the Flow
Cytometer Parameters
Using FC, we extracted the values for FITC, FSC,
SSC and W for each cell. The flow cytometer also
informs on the ‘Height’ (H) and ‘Area’ (A) of the
signals, except for W. We evaluated the correlation
between the H and A signals of FITC, FSC, and SSC
by least-squares fits (Methods) to measure the R
2
of
fitted regression lines, along with the P-value of
statistical significance (Table 1).
In all cases we obtained ‘high’ positive R
2
values
indicating that the fit approximates well the data, in
a positive fashion. Further, all P-values are smaller
than 0.01, from which we conclude that the data is
well explained by a linear least-squares regression fit
between the pairs of variables. As such, from here
onwards, we only use the parameters FITCH, FSCH
and SSCH, along with W.
3.3 RNA Polymerase Numbers as a
Function of Medium Richness
For this, we measured the single-cell fluorescence
intensities of RNAp (FITCH channel) in each
medium. Figure 2 shows the distribution of the
number of cells with given FITCH values for each
medium.
Figure 2: Distribution of the number of cells with given
values of FITCH in each medium, as measured by flow
cytometry: Left: M63; Middle: LB; Right: TB.
To assess if the distributions differ statistically,
we performed Kolmogorov-Smirnov tests (KS-test)
of statistical significance between all pairs of
conditions (the null hypothesis is that the two data
sets belong to the same distribution). In all cases, the
P-value was smaller than 0.05, from which we
conclude that they differ in a statistical sense.
Table 2: Mean and coefficient of variation (CV) of the
distributions of FITCH (proxy for RNAp numbers) in each
medium condition.
Medium Mean(FITCH) CV(FITCH)
M63 2.6x10
3
0.47
LB 2.4x10
3
0.40
TB 2.8x10
3
0.37
To assess the behavioral trend of RNAp levels
with increasing medium richness, we first calculated
the mean and coefficient of variation (CV) of each
distribution. From Table 2, we find that the
CV(FITCH) decreases with medium richness, while
the mean(FITCH) is minimized in LB medium.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
228
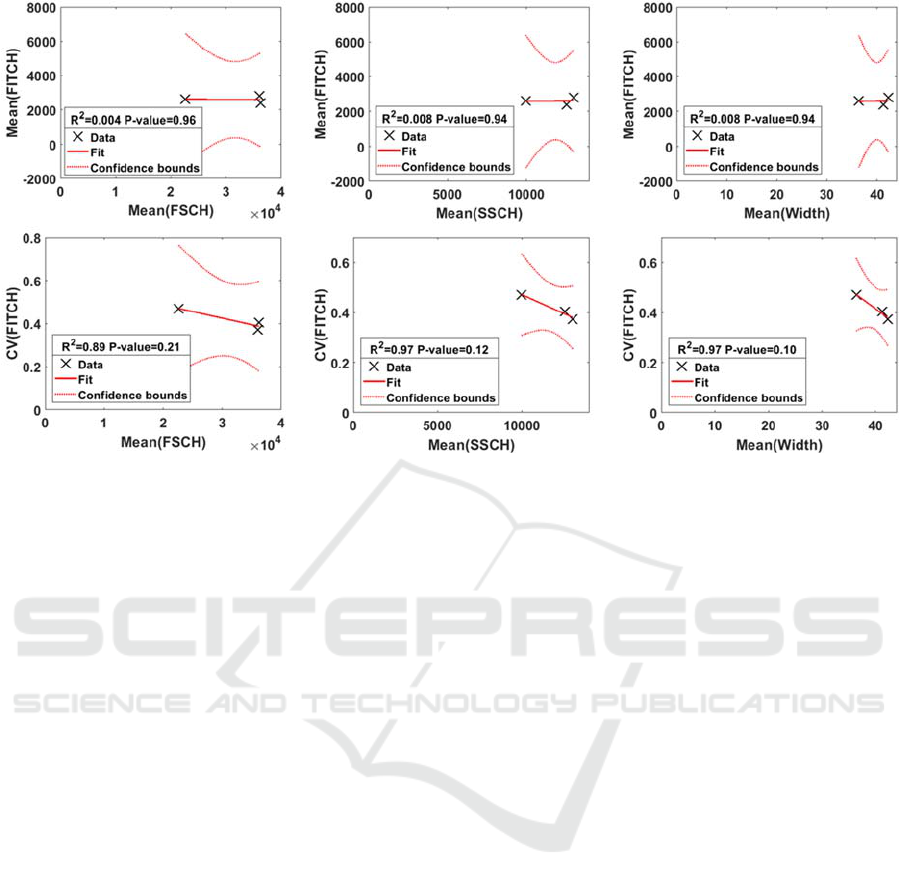
Figure 3: Top: Mean values of FITCH (proxy for RNAp numbers) plotted against the mean values of FSCH, SSCH and
Width (proxies for cell size and composition), respectively, in the three media considered (M63, LB, and TB), along with
the linear least-squares regression fits and confidence intervals. Bottom: coefficient of variation (CV) of FITCH values
plotted against the mean values of FSCH, SSCH and Width, respectively, in the three media considered (M63, LB, and TB),
along with the linear least-squares regression fits and confidence intervals.
3.4 Cell Morphology and Composition
as a Function of Medium Richness
Next, we investigated how the morphology and
composition as seen by parameters obtained by FC
differ, at the population level, with medium richness.
For this, we obtained the mean and CV of FSCH,
SSCH, and W at the single-cell level, in each
medium (Table 3). We find that, in general, the
mean values of FSCH, SSCH, and W increase with
increasing medium richness. Meanwhile, their CV
do not exhibit (linear) relationships with medium
richness.
3.5 Correlation between Cell
Morphology and Composition and
RNAp Levels against Medium
Richness
To validate the above conclusions, we tested for the
occurrence of linear correlations between the mean
values of FSCH, SSCH and W with the mean and
CV of RNAp levels as a function of medium
richness. Figure 3 (Top), shows that there are no
such statistically significant correlations.
Similarly, from Figure 3 (Bottom), there are no
statistically significant negative correlations between
the cell-to-cell variability in RNAp levels and the
mean values of FSCH, SSCH and W. However, if
more conditions were considered (e.g. medium of
intermediate richness between those tested), linear
correlations might become statistically significant.
Thus, we hypothesized that FSCH, SSCH and W,
which differ with medium richness, are negatively
correlated to the cell-to-cell variability in RNAp, but
not to the mean.
3.6 Correlation by Classes between
Cell Morphology and Composition
and RNAp Levels
From the data, it is visible the presence of much cell-
to-cell variability in FSCH, SSCH and W, even
within a given medium condition. This hampers the
ability to detect correlations between these
parameters and RNAp levels.
However, if such correlations exist, they should
become enhanced if, instead of analyzing the data
based on the growth condition, one instead classifies
the cells based on the values of FSCH, SSCH and W
(top panels in Figure 4).
Mean and Variability in RNA Polymerase Numbers Are Correlated to the Mean but Not the Variability in Size and Composition of
Escherichia Coli Cells
229
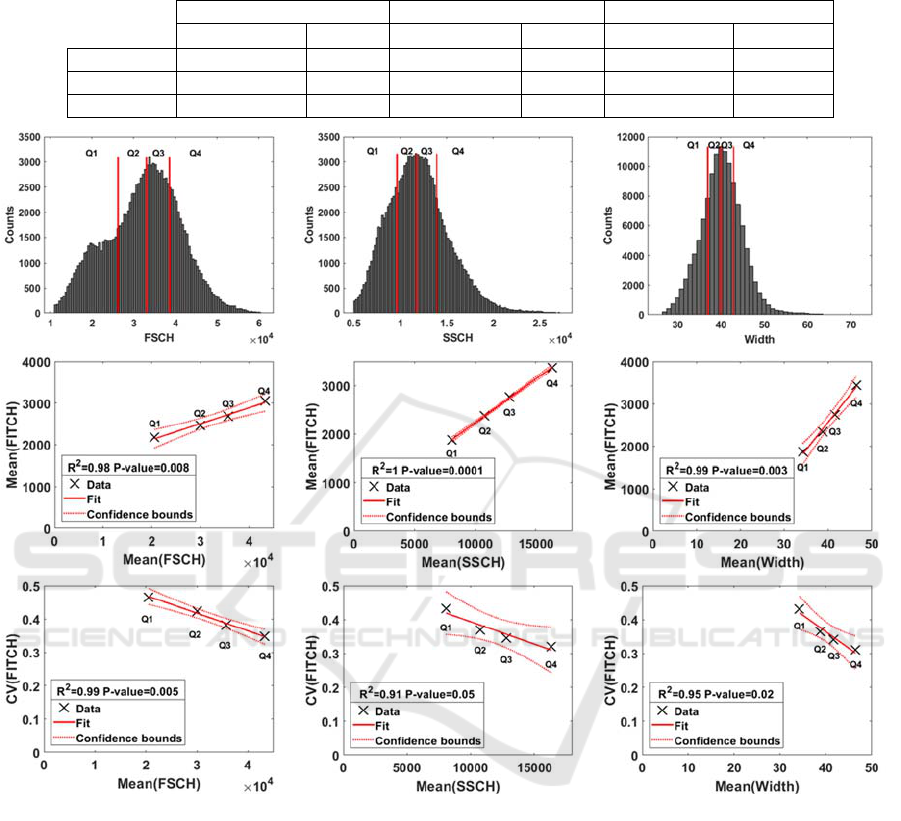
Table 3: Mean and coefficient of variation (CV) of FSCH, SSCH, and Width (proxies for cell size and composition) in each
medium condition.
M63 LB TB
Mean CV Mean CV Mean CV
FSCH 2.26x10
4
0.27 3.62x10
4
0.21 3.61x10
4
0.15
SSCH 9.98x10
3
0.27 1.26x10
4
0.28 1.30x10
4
0.21
W 36.41 0.12 41.30 0.01 42.29 0.09
Figure 4: Top: Distributions of FSCH, SSCH and Width (proxies for cell size and composition) values in individual cells
from all media; Center: Division of the data sets into quartiles and scatter plots Mean(FSCH) and Mean(FITCH, proxy for
RNAp numbers), Mean(SSCH) and Mean(FITCH) , and Mean(Width) and Mean(FITCH); Bottom: Division of the data sets
into quartiles and scatter plots Mean(FSCH) and CV(FITCH), Mean(SSCH) and CV(FITCH) , and Mean(Width) and
CV(FITCH).
We expect that, if the mean values of SSCH,
FSCH, and W can explain the CV(FITCH), then the
linear correlations should be equal or stronger than
when partitioning the data according to the medium.
Further, the P-values should be smaller than 0.01,
implying that the correlations are statistically
significant.
Figure 4 validates this hypothesis, i.e., when
partitioning cells according to the values of SSCH,
FSCH, and W, respectively, one finds strong,
statistically significant, negative linear correlations.
We conclude that the cell-to-cell variability in
RNAp levels decreases for increasing mean values
of FSCH, SSCH, and/or W, which are proxies for
cell size and/or density. Meanwhile, also from
Figure 4, mean RNAp levels increase with mean
values of FSCH, SSCH, and W.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
230
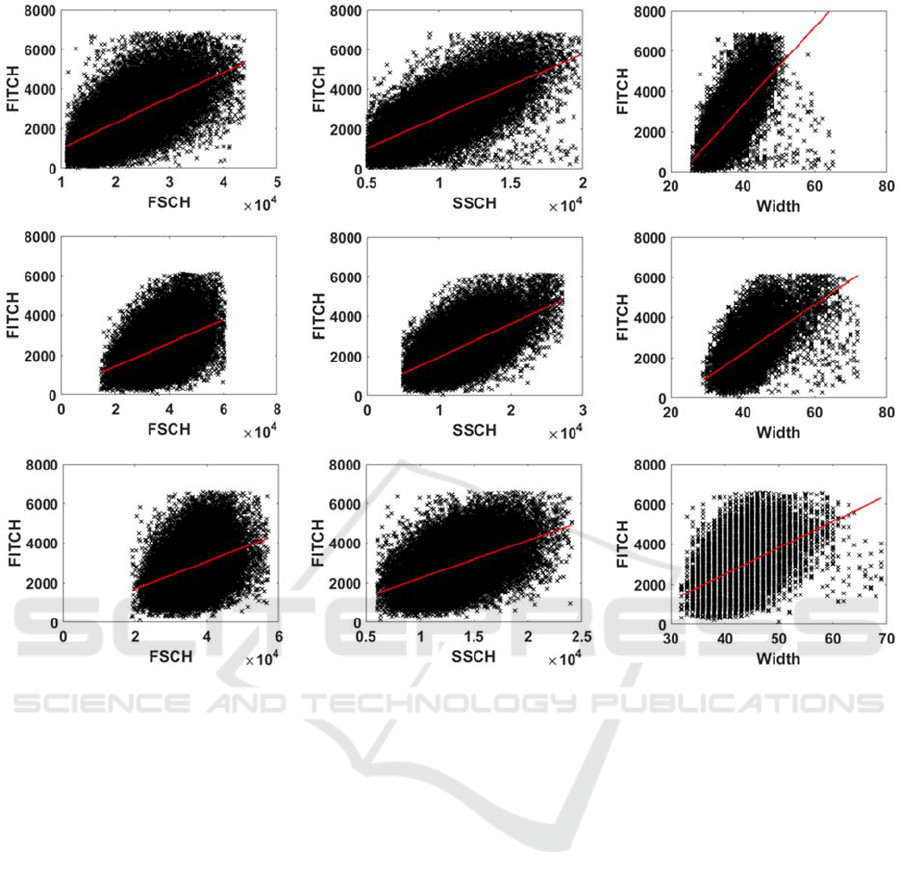
Figure 5: Scatter plots between single-cell values of FITCH (proxy for RNAp numbers) and FSCH, SSCH and Width
(proxies for cell size and composition), respectively, in each medium. Top: M63; Center: LB; Bottom: TB. The solid red
line is the linear least-squares regression fit.
3.7 Correlation by Classes between the
Cell-to-cell Variability in Cell
Morphology and Composition and
in RNAp Levels
We searched for correlations between the cell-to-cell
variability in cell morphology and composition and
the mean and the cell-to-cell variability in RNAp
levels. To obtain classes of cells with differing
variability in these parameters, we made use of
random sampling from the entire set of cells
gathered from all conditions. Namely, for
assembling the values for each class, we randomly
selected 10000 cells and obtained the CV of this set.
This was performed 1000 times. Next, from the
1000 sets, we selected the 10 sets with minimal and
the 10 sets with maximal cell-to-cell variability in
FSCH, SSCH, and W, respectively. We obtained the
CV of the parameter value for each set, and
calculated the average CV of the 10 sets of cells. For
each of these sets, we also obtained the mean and
CV of the RNAp levels of individual cells. As we
found no statistically significantly linear correlation
(R
2
values below 0.15), we conclude that, unlike for
mean values, the cell-to-cell variability in SSCH,
FSCH, and W, cannot explain the mean and cell-to-
cell variability in RNAp numbers.
3.8 Relationship between Cell
Morphology and Composition and
the RNAp Levels at the Single-Cell
Level
Having found a correlation between the mean and
cell-to-cell variability in RNAp levels and the mean
values of FSCH, SSCH, and/or W of cell
populations, we studied whether such correlations
are significant at the single-cell level, i.e. in a
population of cells in the same medium.
Mean and Variability in RNA Polymerase Numbers Are Correlated to the Mean but Not the Variability in Size and Composition of
Escherichia Coli Cells
231
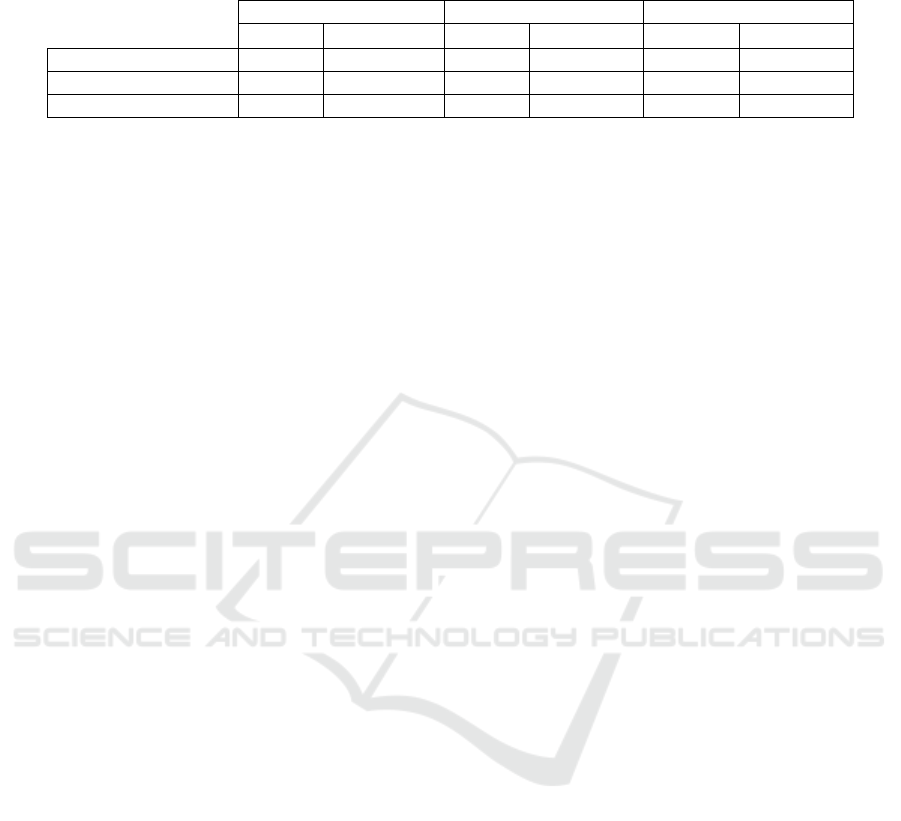
Table 4: Correlation (R
2
) between FITCH (proxy for RNAp numbers) and FSCH, SSCH, and Width (proxies for cell size
and composition) in each medium.
M63 LB TB
R
2
P-value R
2
P-value R
2
P-value
FITCH vs FSCH 0.39 <0.01 0.21 <0.01 0.13 <0.01
FITCH vs SSCH 0.49 <0.01 0.36 <0.01 0.25 <0.01
FITCH vs W 0.47 <0.01 0.34 <0.01 0.24 <0.01
We searched for correlations between single-cell
values of FITCH and the respective values of FSCH,
SSCH and W (Figure 5), by performing fits by linear
regression (least-squares fit method). Also, we
obtained the P-values of statistical significance
(Table 4), by applying F-tests (Methods).
From Figure 5 and Table 4, in all media, the
linear fits are statistically significant, as the P-values
from the least-squares regression fits are smaller
than 0.01 (Table 4). Meanwhile, from the R
2
values,
we find that the goodness of fit decreases for
increasing medium richness.
4 CONCLUSIONS
Our results indicate that the mean and cell-to-cell
variability in RNAp numbers in E. coli cells differs
with parameter values associated to the cell size and
composition, as measured by flow cytometry. In
particular, the mean increases and the variability
decreases as each of these parameter values
increases. At the population level, these changes can
only be detected by classifying cells according to the
values of FSCH, SSCH and Width, respectively.
Analyzing the data at the single-cell level, one also
finds these correlations, being more pronounced in
poor growth medium.
We expect this knowledge to be relevant in
studies of gene expression dynamics in various
media, as the amount of RNAp is a key regulatory
mechanism of transcription dynamics. Namely, our
results suggest that the cell-to-cell variability in gene
expression may differ not only due to intrinsic noise
in gene expression and extrinsic factors, but also due
to the medium-dependence of the mean values of
FSCH, SSCH and Width.
At present, we cannot explain why the cell-to-
cell variability in SSCH, FSCH, and Width are not
correlated to the cell-to-cell variability in RNAp
numbers, while being correlated to the mean RNAp
numbers. An answer to this question may be of
relevance, as the RNAp is a master regulator of gene
expression in bacteria, and the answer may reveal
aspects of how their numbers are regulated. Thus,
the answers should contribute to a better
understanding of the modifications that these
organisms undergo following environmental
changes.
REFERENCES
Arkin, A., Ross, J. and McAdams, H. H. (1998)
‘Stochastic kinetic analysis of developmental pathway
bifurcation in phage λ-infected Escherichia coli cells’,
Genetics, 149(4), pp. 1633–1648. doi: 10.1016/0092-
8674(82)90456-1.
Bratton, B. P., Mooney, R. A. and Weisshaar, J. C. (2011)
‘Spatial distribution and diffusive motion of rna
polymerase in live Escherichia coli’, Journal of
Bacteriology, 193(19), pp. 5138–5146. doi:
10.1128/JB.00198-11.
Browning, D. F. and Busby, S. J. (2016) ‘Local and global
regulation of transcription initiation in bacteria’,
Nature Reviews Microbiology. Nature Publishing
Group, 14(10), pp. 638–650. doi:
10.1038/nrmicro.2016.103.
Cabrera, J. E. and Jin, D. J. (2003) ‘The distribution of
RNA polymerase in Escherichia coli is dynamic and
sensitive to environmental cues’, Molecular
Microbiology, 50(5), pp. 1493–1505. doi:
10.1046/j.1365-2958.2003.03805.x.
Elowitz, M. B. et al. (2002) ‘Stochastic gene expression in
a single cell: Supporting online material’, Science,
297, pp. 1183–1187.
Gillespie, D. T. (1977) ‘Exact stochastic simulation of
coupled chemical reactions’, Journal of Physical
Chemistry, 81(25), pp. 2340–2361. doi:
10.1021/j100540a008.
Kærn, M. et al. (2005) ‘Stochasticity in gene expression:
From theories to phenotypes’, Nature Reviews
Genetics, 6(6), pp. 451–464. doi: 10.1038/nrg1615.
Mäkelä, J., Kandavalli, V. and Ribeiro, A. S. (2017)
‘Rate-limiting steps in transcription dictate sensitivity
to variability in cellular components’, Scientific
Reports, 7(1), pp. 10588. doi: 10.1038/s41598-017-
11257-2.
McClure, W. R. (1980) ‘Rate-limiting steps in RNA chain
initiation.’, Proceedings of the National Academy of
Sciences, 77(10), pp. 5634–5638. doi:
10.1073/pnas.77.10.5634.
McClure, W. R. (1985) ‘Mechanism and Control of
Transcription Initiation in Prokaryotes’, Annual
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
232

Review of Biochemistry, 54(1), pp. 171–204. doi:
10.1146/annurev.bi.54.070185.001131.
Muthukrishnan, A. B. et al. (2014) ‘In vivo transcription
kinetics of a synthetic gene uninvolved in stress-
response pathways in stressed Escherichia coli cells’,
PLoS ONE, 9(9). doi: 10.1371/journal.pone.0109005.
NSM Goncalves et al (2018) Temperature-dependence of
the single-cell kinetics of transcription activation in
Escherichia coli. Physical Biology 15(2):026007.
DOI:10.1088/1478-3975/aa9ddf
Oliveira, S. M. D. et al. (2016) ‘Temperature-Dependent
Model of Multi-step Transcription Initiation in
Escherichia coli Based on Live Single-Cell
Measurements’, PLoS Computational Biology, 12(10).
doi: 10.1371/journal.pcbi.1005174.
Yang, S. et al. (2014) ‘Contribution of RNA polymerase
concentration variation to protein expression noise’,
Nature Communications, 5, pp. 1–9. doi:
10.1038/ncomms5761.
Mean and Variability in RNA Polymerase Numbers Are Correlated to the Mean but Not the Variability in Size and Composition of
Escherichia Coli Cells
233
