
Web-based Interactive Visualization of Medical Images in a Distributed
System
Thiago Moraes
1
, Paulo Amorim
1
, Jorge Silva
1
and Helio Pedrini
2
1
Division of 3D Technologies, Center for Information Technology Renato Archer, 13069-901, Campinas, SP, Brazil
2
Institute of Computing, University of Campinas, 13083-852, Campinas, SP, Brazil
Keywords:
Remote Medical System, Web-based Interactive Visualization, Medical Images, Data Rendering.
Abstract:
Medical images play a crucial role in the diagnosis and treatment of diseases, since they allow the visualization
of patient’s anatomy in a non-invasive way. With the advances of acquisition equipments, medical images
have become increasingly detailed. On the other hand, they have required more computational infrastructure
in terms of processing and memory capabilities, which may not be available in hospitals and clinics with
limited resources. Therefore, a convenient mechanism for providing a more effective health service is through
a remote access system. In this paper, we describe and analyze a distributed system for Web-based interactive
visualization of medical volumes.
1 INTRODUCTION
Data visualization (Pandey et al., 2014, Telea, 2014,
Murray, 2017) is an active research field with appli-
cations in several domains of knowledge, for instance,
medicine, biology, remote sensing, meteorology, en-
tertainment, among others.
The increasing amount of acquired data has be-
come a challenge for the traditional visualization pro-
cess performed locally on a client machine, since it
relies on computationally expensive resources to ma-
nipulate and display data volumes. Thus, the client
machine must have sufficient processing and storage
capacity.
On the other hand, remote data visualization (En-
gel et al., 2000, Evesque et al., 2002, Wessels et al.,
2011, Blazona and Mihajlovic, 2007, Marion and
Jomier, 2012) allows costly operations to be per-
formed on a server with high processing and storage
capacity, resources for providing multi-user collabo-
ration, security mechanisms, as well as efficient ver-
sion control of applications.
The high amount of data produced in the med-
ical field requires the use of efficient image analy-
sis techniques (Amorim et al., 2018b, Amorim et al.,
2013,Moraes et al., 2018,Amorim et al., 2018a,Faza-
naro et al., 2016, Ward et al., 2015, Prince and Links,
2014), which usually involve the intervention of spe-
cialists to allow greater control of the application.
Examples of interactive medical applications include
image segmentation, surgery simulation, virtual and
augmented reality, telemedicine, computer-aided di-
agnosis, among others.
Advances in Web technologies have made it pos-
sible to develop collaborative visualization of med-
ical data on the Internet, improving healthcare ser-
vices. Despite the availability of hardware and soft-
ware support, there are still some challenges in Web-
based medical data visualization.
In this work, we present and analyze a remote sys-
tem for medical data visualization on the Web. From
requests sent to a server, users are able to manipu-
late and visualize large volumes of data quickly and
directly in the Web browsing environment. Several
benefits are offered to users from the proposed sys-
tem. Web browser-based rendering requires no spe-
cial programs to be installed on the client machine.
As main contributions of the paper, the server is
responsible for performing the operations requested
by the users, which dispenses the client machine from
expensive requirements in terms of memory and pro-
cessing. In addition, viewing results become available
from any device with Internet access. The server ma-
chine is a distributed system in order to provide capa-
bilities of performance, expansibility and reliability.
The proposed solution uses only open and free pack-
ages.
This text is organized as follows. Section 2 briefly
presents concepts and approaches related to the topic
under investigation. Section 3 describes the method-
346
Moraes, T., Amorim, P., Silva, J. and Pedrini, H.
Web-based Interactive Visualization of Medical Images in a Distributed System.
DOI: 10.5220/0007626103460353
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 346-353
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

ology proposed to develop a remote medical data vi-
sualization system on the Web. Experimental results
are presented and evaluated in Section 4. Concluding
remarks and directions for future work are outlined in
Section 5.
2 BACKGROUND
Medical volume rendering consists of a set of tech-
niques for displaying a two-dimensional (2D) of a
three-dimensional data set, acquired by a medical
scanner such as Magnetic Resonance Imaging (MRI)
and Computed Tomography (CT).
The advances and diffusion of the Internet have
leveraged the interest in developing tools for analy-
sis and processing of medical images in an Internet
browser.
Virtual Reality Modeling Language
(VRML) (VRML, 2012) was one of the first
proposals for modeling virtual reality in Web envi-
ronments, allowing users connected to the Internet
to interactively view data volumes and transmit
files. However, VRML has not become a Web
standard supported by the majority of the browsers,
so it is necessary to install a plugin in the browser.
Current leading browsers have adopted two important
standards: WebGL and WebSocket.
WebGL (WebGL, 2017) is the OpenGL ES stan-
dard for browsers. It allows the creation of OpenGL
contexts in browser windows and programming with
the Javascript language to create three-dimensional
interactive environments.
WebSocket (Fette and Melnikov, 2011) is a net-
work protocol that allows two-way communication
between client and server. Unlike Hypertext Transfer
Protocol (HTTP), communication between both ma-
chines (client and server) remains active, which facil-
itates the transmission and development of content in
real time.
Distributed systems (Tanenbaum and Van Steen,
2007) refers to an architecture in which computing
nodes in a network communicate via messages to co-
ordinate themselves in order to achieve a common
goal. An interesting feature of this system is that fail-
ure of a node should not affect the entire system.
Visualization Toolkit (Schroeder et al., 2004,
VTK, 2018) and Three.js (Dirksen, 2013) are some
examples of libraries used to create and visualize data
in a Web browser.
Publisher-Subscribe (PubSub) (Ion et al., 2010) is
an asynchronous message passing model, where pub-
lishers and subscribers exchange messages without
knowledge of each other. Messages are called events.
In the PubSub model, subscribers signalize to the bro-
ker as interested in a particular event. The publisher
sends messages (events) to the broker, which is re-
sponsible for delivering the message to the subscriber
registered in the event. Redis (Carlson, 2013, Redis-
Lab, 2018) is an open source in-memory database that
can be used as broker, since it implements the Pub-
Sub model. Redis allows the broker to be distributed
across multiple nodes.
Some initiatives to develop data visualization sys-
tems on the Web have been made available in the liter-
ature (Kokelj et al., 2018, Rego and Koes, 2015, Kas-
par et al., 2013,Sherif et al., 2015,Marion and Jomier,
2012,Mahmoudi et al., 2010,Mwalongo et al., 2015).
3 REMOTE SYSTEM FOR
MEDICAL DATA
VISUALIZATION
In this section, we describe the developed remote
medical data visualization system on the Web. Fig-
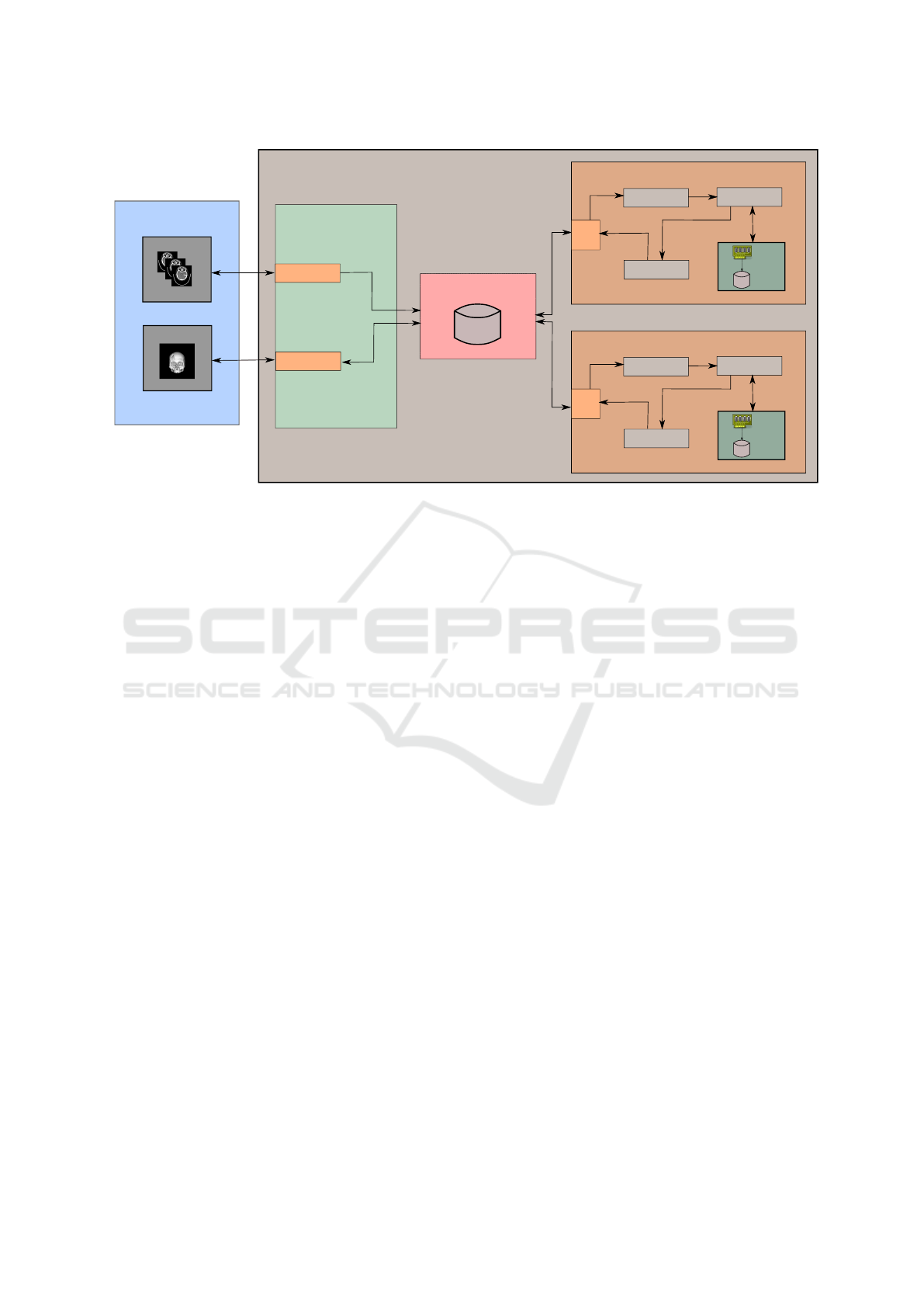
ure 1 illustrates the main components of the proposed
remote system for medical data visualization. The
system is composed of three different modules:
• Web Server: where the HTTP (Hypertext Trans-
fer Protocol) (Fielding et al., 1999) and Web-
Socket (Fette and Melnikov, 2011) requests are
received from the clients. The Flask frame-
work (Ronacher, 2018) is used to generate
and send HTML (HyperText Markup Language)
pages to the client from HTTP requests. Flask-
SockeIO (Grinberg, 2018) is responsible for re-
ceiving and responding to WebSocket requests.
• Redis Server: Redis (Carlson, 2013, RedisLab,
2018) is an in-memory database that can be used
as broker since it implements the PubSub model.
Redis allows the broker to be distributed across
multiple nodes, that is, more than one Redis server
can be used.
• Processing Server: one or more processing nodes.
It was implemented in Python programming lan-
guage.
All these modules may reside either on the same
machine or on different machines interconnected by a
data network. Redis performs load balancing to avoid
processing overhead on only one of the processing
servers. The client is a Web browser implemented in
HTML and Javascript languages.
In this architecture, the processing servers sub-
scribe to the broker (Redis server) to receive events.
Web-based Interactive Visualization of Medical Images in a Distributed System
347
Web Server
HTTP and WebSocket
Flask-SocketIO
Flask
Client (browser)
HTML + Javascript + WebGL
Slices
3D Surface
3D Canvas
3D Canvas
Redis Server
DICOM
files
NumPy
VTK
GDCM
MMAP
Processing Server 0
Redis
Client
...
Remote System
NumPy
VTK
GDCM
MMAP
Processing Server n
Redis
Client
Figure 1: Diagram that illustrates the main stages of the proposed remote system for medical data visualization.
Examples of events include: 3D reconstruction, seg-
mentation, mesh generation, and mesh rendering. The
Web server receives client requests (via HTTP or
WebSocket) and then sends an event to Redis with the
data received from the client.
Redis stores the data in memory (or disk, if the
data is too large) and contacts one of the processing
servers registered in that event. The processing server
accesses event data and performs processing. Af-
ter performing the processing, the processing server
sends a Redis event with the resulting data. The Web
server subscribes to an event to receive these results.
After receiving the results of the processing, the Web
server sends them to the client.
Events in Redis are identified by a name (string).
In order for the processing server to receive events
related to the images in its possession, it subscribes
to dynamically generated events. The same occurs
with the Web server. This is necessary to not oc-
cur from the Web server sending the result to other
client than the one that requested the result. This
event name is generated by concatenating the oper-
ation to be performed with the image identifier. For
instance, the segmentation operation for image 42
may have the event name segment_image_00042,
whereas the Web server expects the result with the
event result_segmentation_image_00042.
The workflow of the methodology starts when the
client sends medical images (for instance, Computed
Tomography, Magnetic Resonance Imaging, Micro-
tomography) in the format DICOM (ACR/NEMA,
2018) to the distributed system via HTTP.
In the distributed system, the images are received
by the Web server, which sends these images to one of
the processing servers through a Redis event. When
the event arrives at the processing server, it retrieves
the images from the Redis server and removes those
Redis images. From then on, all processing in these
images is performed on that same processing server.
This is done to avoid leaving the images stored on the
Redis server.
On the processing server, the GDCM li-
brary (GDCM, 2018) is used to rebuild the im-
ages on a volume. The resulting volume is con-
verted to a volumetric array using the numeric li-
brary Numpy (Oliphant, 2007) in conjunction with
the VTK (Visualization Toolkit) library (Schroeder
et al., 2004,VTK, 2018). The volumetric array is then
written to a disk file. The file is mapped to mem-
ory via MMAP (Memory Map) (MMAP, 2018) using
Numpy. Thus, only the portions of the data required
for processing are kept in main memory and not the
entire volume, which reduces the amount of memory
required in the processing server.
After the data preparation (both two-dimensional
and three-dimensional data) for visualization purpose,
all the processing of subsequent steps is performed in
the distributed system, where the client needs only to
request data from the system and interpret them. All
the available steps are listed as follows:
1. 3D Image Slice Visualization Tool: this stage aims
to allow the visualization of the slices in the ax-
ial, sagittal and coronal orientations. It starts with
the request, by the client, using the HTTP pro-
tocol, of a given slice and orientation of interest.
The Web server receives the request and sends an
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
348

event to Redis. The processing server with the
corresponding image receives the event with the
required data. The processing server retrieves the
slice of interest in the file containing the volumet-
ric array. Slice retrieval is done through file access
with Numpy and MMAP. The processing server
sends an event to Redis with the desired slice. The
Web server subscribes to the corresponding event
to receive the image. After receiving the image, it
sends the image to the client. In possession of the
image, the client renders it on the browser screen.
Rendering is done by mapping the image in a
plane as a texture. WebGL (WebGL, 2017) is used
for rendering through JavaScript three.js (Cabello,
2018) library.
2. Segmentation Tool: segmentation is done through
the double thresholding technique. In this ap-
proach, voxels are selected with gray levels be-
tween T
min
and T
max
. The values of T
min
and T
max
are parameters controlled by the user through a
graphical interface on the client. T
min
and T
max
are
sent to the distributed system via HTTP. The Web
server receives the request with the segmentation
data and sends an event. This event is received by
the processing server with the corresponding im-
age to be segmented. It segments the images and
sends the response through an event to the Web
server. It sends a representation of this segmenta-
tion to the client via HTTP, which is a mask (in
green color) superimposed on the voxels of the
slice that are within the range of the threshold.
3. Mesh Generation Tool: after segmentation, the
user can generate the three-dimensional surface,
which is produced by the processing server us-
ing the marching cubes algorithm (Lorensen and
Cline, 1987), implemented in the VTK library. A
simplified version, containing fewer polygons of
the three-dimensional surface, is also generated.
The simplified surface is created using the deci-
mation algorithm (Garland and Heckbert, 1997),
implemented in the VTK library. The process-
ing server sends the simplified surface to the Web
server, which sends it to the client using the Web-
Socket protocol.
4. 3D Triangle Meshes Visualization Tool: with the
simplified surface, the client renders it using We-
bGL. Rendering of the simplified version oc-
curs during user interaction with the visualiza-
tion. After the interaction, the client sends a mes-
sage requesting the rendering of the entire three-
dimensional surface. This request is made using
WebSocket and contains camera information (po-
sition, focus, and vector up that indicates the top
side of the camera). This request arrives at the
Web server, which sends the camera data to the
processing server, via an event. The processing
server renders the entire surface offscreen (Paul,
1997), according to the information sent by the
client. The result is an image, which is sent to the
Web server via an event. The Web server sends
the client via WebSocket. The client then renders
this image as plane texture with same dimensions
as the renderer.
4 RESULTS
This section presents the results obtained with the im-
plementation of the proposed methodology. Three
server-side machines were used: (i) 1 server with Re-
dis, (ii) 1 Web server with the proposed system and
one of the processing nodes, (iii) 1 processing node.
On the client side, browsers were Mozilla Fire-
fox (Desktop), Chrome (Android, Mobile) and Safari
(iOS, Mobile). The only requirements on the client
side are the availability of WebGL, WebSocket and
Javascript.
Figure 2 illustrates the view of slices in the axial,
coronal and sagittal orientations. Below each slice,
there are sliders to change the slice to be displayed.
In the lower right corner, it is possible to observe the
segmentation tool based on thresholding. The thresh-
old values are changed by dragging the sliders or typ-
ing the values directly. In the slices, it is possible to
notice the mask (in green) superimposed on the slice
indicating the segmentation.
Figure 3 shows the visualization of a three-
dimensional surface. The visualization is done in two
parts: the rendering of the simplified mesh, which is
performed on the client, and the complete rendering,
which is performed by the server. Simplified ren-
dering is performed while the user interacts with the
mesh. Figure 3a illustrates this rendering. Figure 3b
shows the result of rendering the complete surface,
which is performed after the user interaction.
The proposed architecture proved to be very flex-
ible by allowing the tool to be used on different mo-
bile systems, such as Android and iOS. This makes
it possible to use the system in non-desktop environ-
ments, which could occur during a doctor’s visit to a
patient’s bed. Figure 4 illustrates the surface render-
ing interface on Android 6.0 and iOS 10.3 systems.
Web-based Interactive Visualization of Medical Images in a Distributed System
349

Figure 2: System interface used to visualize the slices in the axial, coronal and sagittal orientations.
(a) simplified surface (b) complete surface
Figure 3: Surface rendering on a Web browser.
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
350

(a) Android (b) iOS
Figure 4: Result of surface rendering on a smartphone Android version 6.0 and iOS 10.3.
5 CONCLUSIONS AND FUTURE
WORK
The distributed system for medical image visualiza-
tion via Web was implemented and proved to be very
useful. This makes it possible to centralize the pro-
cessing and offer such computing resources to hospi-
tals and clinics that do not have much computational
power.
The possibility of accessing the system via mobile
browser will allow access to the rendering system on
smartphones, which may be useful for physicians and
dentists when a desktop computer is not available.
Directions for future work include improvements
in triangle mesh generation, automated tools for cor-
recting mesh problems (such as holes and face normal
reversals), and other segmentation tools, for instance,
watershed and region growing. We also intend to in-
corporate the developed system into the open-source
InVesalius (InVesalius, 2018, Amorim et al., 2015).
ACKNOWLEDGMENTS
The authors thank FAPESP (grants #2014/12236-1
and #2017/12646-3), CNPq (grant #305169/2015-7)
and CAPES for the financial support.
REFERENCES
ACR/NEMA, D. (2018). Digital Imaging and Communica-
tions in Medicine. http://dicom.nema.org/.
Amorim, P., Moraes, T., Silva, J., and Pedrini, H. (2013).
An Out-of-Core Volume Rendering Architecture. In
IV ECCOMAS Thematic Conference on Computa-
tional Vision and Medical Image Processing, pages
173–179, Funchal, Portugal. Taylor & Francis Group,
CRC Press/Balkema.
Amorim, P., Moraes, T., Silva, J., and Pedrini, H. (2015).
InVesalius: An Interactive Rendering Framework for
Health Care Support. In 11th International Sympo-
sium on Visual Computing, volume LNCS 9474, pages
45–54, Las Vegas, NV, USA. Springer-Verlag.
Amorim, P., Moraes, T., Silva, J., and Pedrini, H. (2018a).
3D Adaptive Histogram Equalization Method for
Medical Volumes. In 13th International Conference
Web-based Interactive Visualization of Medical Images in a Distributed System
351

on Computer Vision Theory and Applications, pages
363–370, Funchal, Madeira, Portugal.
Amorim, P., Moraes, T., Silva, J., and Pedrini, H. (2018b).
Out-of-Core Rendering of Large Volumetric Data Sets
at Multiple Levels of Detail. In Multi-Modality Imag-
ing, pages 191–215. Springer International Publish-
ing.
Blazona, B. and Mihajlovic, Z. (2007). Visualization Ser-
vice Based on Web Services. Journal of Commputing
and Information Technology, 4:339–345.
Cabello, R. (2018). three.js - JavaScript 3D Library.
http://mrdoob.github.com/three.js/.
Carlson, J. L. (2013). Redis in Action. Manning Publica-
tions Co., Greenwich, CT, USA.
Dirksen, J. (2013). Learning Three.js: The JavaScript 3D
Library for WebGL. Packt Publishing Ltd.
Engel, K., Ertl, T., Hastreiter, P., Tomandl, B., and Eber-
hardt, K. (2000). Combining Local and Remote Visu-
alization Techniques for Interactive Volume Render-
ing in Medical Applications. In Conference on Vi-
sualization, pages 449–452. IEEE Computer Society
Press.
Evesque, F., Gerlach, S., and Hersch, R. (2002). Building
3D Anatomical Scenes on the Web. Journal of Visu-
alization and Computer Animation, 13(1):43–52.
Fazanaro, D., Amorim, P., Moraes, T., Silva, J., and Pedrini,
H. (2016). NURBS Parameterization for Medical Sur-
face Reconstruction. Applied Mathematics, 7(2):137–
144.
Fette, I. and Melnikov, A. (2011). The WebSocket Protocol.
RFC 6455 (Proposed Standard).
Fielding, R., Gettys, J., Mogul, J., Frystyk, H., Masinter,
L., Leach, P., and Berners-Lee, T. (1999). Hypertext
Transfer Protocol – HTTP/1.1. RFC 2616 (Draft Stan-
dard). http://www.ietf.org/rfc/rfc2616.txt.
Garland, M. and Heckbert, P. S. (1997). Surface Simpli-
fication Using Quadric Error Metrics. In 24th An-
nual Conference on Computer Graphics and Interac-
tive Techniques, pages 209–216, Los Angeles, CA,
USA.
GDCM (2018). Grassroots DICOM Library.
http://sourceforge.net/projects/gdcm/.
Grinberg, M. (2018). Flask SocketIO. https://flask-
socketio.readthedocs.io/en/latest/.
InVesalius (2018). Open Source Software for Reconstruc-
tion of Computed Tomography and Magnetic Reso-
nance Images. http://www.cti.gov.br/invesalius/.
Ion, M., Russello, G., and Crispo, B. (2010). Sup-
porting Publication and Subscription Confidentiality
in Pub/Sub Networks. In International Conference
on Security and Privacy in Communication Systems,
pages 272–289, Singapore, Singapore. Springer.
Kaspar, M., Parsad, N., and Silverstein, J. (2013). An Opti-
mized Web-based Approach for Collaborative Stereo-
scopic Medical Visualization. Journal of the Ameri-
can Medical Informatics Association, 20(3):535–543.
Kokelj, Z., Bohaka, C., and Marolt, M. (2018). A web-
based virtual reality environment for medical visual-
ization. In 41st International Convention on Informa-
tion and Communication Technology, Electronics and
Microelectronics, pages 0299–0302.
Lorensen, W. E. and Cline, H. E. (1987). Marching
Cubes: A High Resolution 3D Surface Construction
Algorithm. In 14th Annual Conference on Computer
Graphics and Interactive Techniques, pages 163–169,
New York, NY, USA.
Mahmoudi, S. E., Akhondi-Asl, A., Rahmani, R., Faghih-
Roohi, S., Taimouri, V., Sabouri, A., and Soltanian-
Zadeh, H. (2010). Web-based Interactive 2D/3D Med-
ical Image Processing and Visualization Software.
Computer Methods and Programs in Biomedicine,
98(2):172–182.
Marion, C. and Jomier, J. (2012). Real-Time Collaborative
Scientific WebGL Visualization with WebSocket. In
17th International Conference on 3D Web Technology,
pages 47–50, Los Angeles, CA, USA.
MMAP (2018). Memory-Mapped
I/O. http://www.kernel.org/doc/man-
pages/online/pages/man2/mmap.2.html.
Moraes, T., Amorim, P., Silva, J., and Pedrini, H. (2018).
3D Lanczos Interpolation for Medical Volumes. In
15th International Symposium on Computer Methods
in Biomechanics and Biomedical Engineering, pages
1–10, Lisbon, Portugal.
Murray, S. (2017). Interactive Data Visualization for the
Web: An Introduction to Designing with. O’Reilly
Media, Inc.
Mwalongo, F., Krone, M., Becher, M., Reina, G., and Ertl,
T. (2015). Remote Visualization of Dynamic Molecu-
lar Data using WebGL. In 20th International Confer-
ence on 3D Web Technology, pages 115–122.
Oliphant, T. E. (2007). Python for Scientific Computing.
Computing in Science & Engineering, 9(3):10–20.
Pandey, A. V., Manivannan, A., Nov, O., Satterthwaite,
M., and Bertini, E. (2014). The Persuasive Power of
Data Visualization. IEEE Transactions on Visualiza-
tion and Computer Graphics, 20(12):2211–2220.
Paul, B. (1997). OpenGL/Mesa Off-Screen Rendering.
In ACM SIGGRAPH Conference, Los Angeles, CA,
USA.
Prince, J. and Links, J. (2014). Medical Imaging Signals
and Systems. Pearson Education.
RedisLab (2018). Redis. https://redis.io/.
Rego, N. and Koes, D. (2015). 3Dmol.js: Molecular Visu-
alization with WebGL. Bioinformatics, 31(8):1322–
1324.
Ronacher, A. (2018). Flask. http://flask.pocoo.org/.
Schroeder, W., Martin, K., Martin, K. W., and Lorensen, B.
(2004). The Visualization Toolkit. Prentice Hall PTR.
Sherif, T., Kassis, N., Rousseau, M.-T., Adalat, R., and
Evans, A. (2015). Brainbrowser: Distributed, Web-
based Neurological Data Visualization. Frontiers in
Neuroinformatic, 8:1–10.
Tanenbaum, A. S. and Van Steen, M. (2007). Distributed
Systems: Principles and Paradigms. Prentice-Hall.
Telea, A. C. (2014). Data Visualization: Principles and
Practice. CRC Press.
VRML (2012). Virtual Reality Modeling Language.
http://www.vrml.org/.
VTK (2018). Visualization Toolkit. http://www.vtk.org/.
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
352

Ward, M. O., Grinstein, G., and Keim, D. (2015). Interac-
tive Data Visualization: Foundations, Techniques, and
Applications. AK Peters/CRC Press.
WebGL (2017). OpenGL ES 2.0 for the Web.
http://www.khronos.org/webgl/.
Wessels, A., Purvis, M., Jackson, J., and Rahman, S. (2011).
Remote Data Visualization through WebSockets. In
Eighth International Conference on Information Tech-
nology: New Generations, pages 1050–1051.
Web-based Interactive Visualization of Medical Images in a Distributed System
353
