
Accurate Plant Modeling based on the Real Light Incidence
J. M. Jurado, J. L. C
´
ardenas, C. J. Ogayar, L. Ortega and F. R. Feito
Computer Graphics and Geomatics Group, University of Ja
´
en, Spain
Keywords:
3D Plant Reconstruction, Spectral Reflectance, Image Processing, Procedural Modeling.
Abstract:
In this paper, we propose a framework for accurate plant modeling constrained to actual plant-light interaction
along a time-interval. To this end, several plant models have been generated by using data from different
sources such as LiDAR scanning, optical cameras and multispectral sensors. In contrast to previous approaches
that mostly focus on realistic rendering purposes, the main objective of our method is to improve the multi-
view stereo reconstruction of plant structures and the prediction of the growth of existing plants according to
the influence of real light incidence. Our experimental results are oriented to olive trees, which are formed by
many thin branches and dense foliage. Plant reconstruction is a challenging task due to self-occlusion. Our
approach is based on inverse modeling to generate a parametric model which describes how plants evolve in a
time interval by considering the surrounding environment. A multispectral sensor has been used to characterize
input plant models from reflectance values for each narrow-band. We propose the fusion of heterogeneous data
to achieve a more accurate modeling of plant structure and the prediction of the branching fate.
1 INTRODUCTION
Realistic plant modeling is a well known topic of
research in Computer Graphics. Its applications
are mainly visualization and virtual reality. Other
disciplines have also included plant reconstruction
as a research objective, such as remote sensing
(Prusinkiewicz, 2004) and biology (Omasa et al.,
2006). Many plant modeling approaches have been
developed over the last years. However, the dynamic
plant behaviour, by considering the surrounding envi-
ronment, remains a non-trivial and challenging task.
Procedural modeling approaches can efficiently
synthesize the branching structure of existing real-
world plant from a set of rule-based system (Bene
ˇ
s
et al., 2011) and (Guo et al., 2018). In general,
these methods are aimed to obtain visually accept-
able results for rendering and simulation purposes,
and therefore they are not directly oriented for 3D
modeling real-world vegetation.
Geometry-based methods may also accurately re-
construct the skeletal structure of a tree, but the fo-
liage is difficult to recreate (Xu et al., 2007; Livny
et al., 2010). One of the most promising image-based
methods is Structure-from-Motion (SfM), which is
widely used to generate 3D point clouds from mul-
tiple overlapping images (Lou et al., 2014). However,
this method arises some limitations, and the recon-
struction of complex plant structures is prone to er-
rors. On the other hand, other approaches may effec-
tively reconstruct real plants using an inverse proce-
dural method (Stava et al., 2010). In addition to ge-
ometrical data, some methods also regard additional
environmental factors, such as light incidence (Stava
et al., 2014), which influences the growth of the plant.
However, these estimations are based on a probabilis-
tic model, and not measured from the actual plant.
In this paper, we propose a several improvements
for plant modeling constrained by the real plant-light
interaction along a time interval. We aim to fuse
the plant static reconstruction and inverse procedu-
ral modeling for plant growth prediction. In contrast
to previous approaches that mostly focus on realistic
rendering purposes, the main objective of our method
is to monitor and predict the growth of real plants.
We have tested our approach with olive trees, which
are formed by a complex crown structure with many
self-hidden branches and leaves. An inverse modeling
process is applied for generating the parametric model
to describe several botanic features. To this end, mul-
tispectral images are used to extract reflectance in-
dices for each narrow-band to estimate the plant vigor
and predict its next growth. A semantic classification
of the plant shape is carried out by considering how
plants reflect the light energy.
The paper is organized as follows. Firstly, a re-
view of previous work on plant modeling is presented
(Section 2) and we provide the general overview of
360
Jurado, J., Cárdenas, J., Ogayar, C., Ortega, L. and Feito, F.
Accurate Plant Modeling based on the Real Light Incidence.
DOI: 10.5220/0007686803600366
In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2019), pages 360-366
ISBN: 978-989-758-354-4
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

our method (Section 3). In the following, experimen-
tal results are shown which are obtained from LiDAR
and multiple images (Section 4) and the fusion of
multispectral data to estimate the real light incidence
to the plant model (Section 5). Finally, we discuss
the contributions of the paper and the main topics for
further research (Section 6).
2 RELATED WORK
Plant modeling is a classic topic of research in Com-
puter Graphics. However, the main efforts have been
made for visualization and virtual reality purposes. In
this work, we apply plant modeling for the monitor-
ing and prediction of the evolution of actual plants
and bushes from extensive plantations. According
to the literature, plant modeling approaches may be
mainly classified into three categories: reconstruction
from existing real-world plants, interactive modeling
and procedural or rule-based systems, such as fractals
(Oppenheimer, 1986) and L-Systems (Prusinkiewicz,
1986). Neither geometry-based nor interactive meth-
ods for tree shape reconstruction takes into account
any environmental effects. On the other hand, proce-
dural methods are capable to generate dynamic plant
models whose evolution can be affected by chang-
ing conditions of the environment (Guo et al., 2018).
There are two main methods which may be used for
modeling existing real-world plants: plant static re-
construction and procedural modeling.
Plant Static Reconstruction. Geometry-based meth-
ods mainly depend on the input data, e.g., the image
quality, environmental light during the capture pro-
cess, the shape and texture of the target plant. In
this way, 3D laser scanning can precisely reconstruct
branching plants (Omasa et al., 2006) and can be used
for the automatic reconstruction of the plant skeleton
without overlapped trees segmentation (Livny et al.,
2010). However, this technique implies some draw-
backs as the sensitivity to occlusion, high device cost,
and tedious application in many complex scenes, es-
pecially terrestrial laser scanning (TLS). As a solution
of this problem the airborne LiDAR means a more
efficient solution for scanning extensive plantations
with an ever increasing precision.
On the other hand, multi-view stereo reconstruc-
tion is another category of methods which can be suc-
cessfully used for tree reconstruction. It is based on a
feature-matching process between multitude overlap-
ping images. In this scope, the SfM method is com-
monly applied to generate a 3D point cloud of a plant
by using several images (Quan et al., 2006). This
technique overcomes some problems mentioned be-
fore, although it may not perform efficiently on com-
plex and heterogeneous surfaces where the detection
of key features is more complex. Moreover, this algo-
rithm is based on the scale-invariant feature transform
(SIFT) which may cause errors for the reconstruc-
tion of complex plant models (Lowe, 2004). Conse-
quently, multi-view based realistic tree modeling with
botanical features still pose several limitations. How-
ever, these problems might be partially solved by gen-
erating depth maps for each view, and fusing them
into a dense point cloud reconstruction (Guo et al.,
2018).
Procedural Modeling. Early approaches of plant
modeling focus on the generation of repetitive pat-
terns as fractals or L-Systems (Deussen et al., 1998).
Instead of modeling directly from generative rules,
guided procedural methods have been proposed to
simulate the plant structure from an accurate point
cloud based representation of its branching struc-
ture. Bennes et al. (Bene
ˇ
s et al., 2011) introduce
guided procedural modeling with several geometric
constraints. Nowadays, the reconstruction from point
clouds has received considerable attention (Berger
et al., 2017). In this way, procedural models can
be transformed into certain shapes depending on the
point cloud input that are acquired with static plant
modeling. Modeling trees according to desired shapes
is important for many applications. However, proce-
dural methods have some drawbacks due to the com-
plexity to determine a valid set of input parameters,
such as branching angle, apical and lateral light ef-
fects, pruning factors, growth rate, etc. These features
are very important for the structure of young trees but
they become less relevant for mature plants. Stava et
al. (Stava et al., 2014) proposed an inverse procedu-
ral modeling method for trees, based on a novel para-
metric model that uses Monte Carlo Markov Chains
(MCMC) for calculating the optimal set of procedu-
ral parameters. However, all these approaches gen-
erate static branching structures rather than modeling
dynamic plant behavior.
Procedural methods are capable of generating tree
models by explicitly considering the environment.
Recent approaches are focused on providing more ef-
ficient ways for plant-environment interaction model-
ing. Palubicki et al. (Palubicki et al., 2009) optimize
branch distribution with space colonization method
(Runions et al., 2007) and local competition for light
resources. In each iteration, the space surrounding of
each bud and the optimal direction of shoot growth
are calculated. Both values are needed to create a tree
structure by simulating real environmental properties.
Later, Lei Yi et al. (Yi et al., 2015) demonstrates the
sensitivity of branching distribution to ambient light
Accurate Plant Modeling based on the Real Light Incidence
361
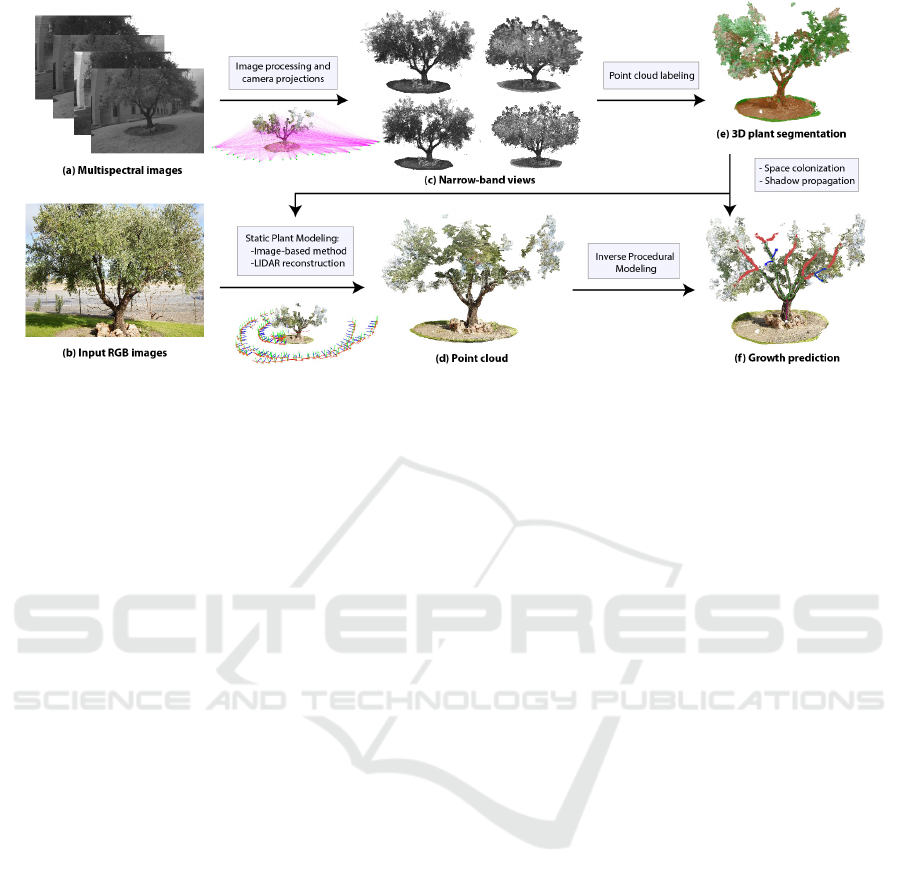
Figure 1: Overview of the method for olive tree reconstruction from the fusion of multispectral data.
and how each branch influences to others.
In addition, an important research topic in Ecol-
ogy and Biology focuses on plant phenotyping and
measuring leaf chlorophyll concentration, in order to
study each growing stage of plants from image-based
remote sensing (Moran et al., 1997). In contrast
to the simply visual detection of diseases, narrow-
band sensors are commonly used to measure plant
changes considering how the vegetation interacts with
the ambient light of the surrounding environment
(Candiago et al., 2015). Unfavorable plant grow-
ing results in morphological, physiological and bio-
chemical changes that are determined by the quan-
tity of absorbed or reflected light. Leaf spectral re-
flectance provides multiple key features for assessing
plant health; however, leaves typically have a low re-
flectance in the visible spectral due to the high chloro-
phyll absorption (Pe
˜
nuelas and Filella, 1998). Mul-
tispectral sensors capture several spectral bands to
detect many properties, such as drought stress, heat
stress, nutrient content and plant biomass. Current ap-
proaches mostly provide a 2D-based analysis for plant
diseases detection (Thomas et al., 2018), image-based
vegetation segmentation (Suh et al., 2018), plant
phenotyping through ground-based sensors (Sankaran
et al., 2010) and LiDAR and hyperspectral remotely
sensed data (Hakkenberg et al., 2018). Although
many of these methods have certain capabilities to de-
scribe growth behaviour, it is quite difficult to acquire
several plant traits which are not directly visible from
remote sensing imagery. This issue might be over-
come through a faithful 3D modeling of the real plant
for a readily comprehensive assessment of complex
plant features (Klodt and Cremers, 2014).
3 OVERVIEW
In this paper, we propose a method for dynamic plant
modeling constrained to actual plant-light interaction
along a time-interval. The incident light is obtained
from several spectral bands and used to improve the
plant static modeling. The main goal is to generate
accurate plant models for monitoring extensive plan-
tations using several 3D capture techniques, working
at different levels of detail. In this work, we focus
on the reconstruction and characterization of a sin-
gle tree model. An inverse modeling process gen-
erates the parametric model from multispectral data
which provide several features to describe the plant
vigor such as the reflectance index, chlorophyll fluo-
rescence or leaves temperature. The generation of the
tree structure by procedural methods is constrained by
several constraints related to the plant status. We also
consider time-lapse data for analyzing the evolution
of the plant along a period. In addition, multispectral
data may be the support for performing reasonably re-
alistic predictions.
The flow diagram of our framework illustrates
the overall process (Figure 1). The first step of our
method is the accurate point cloud reconstruction of
each plant at a detailed level. The first step of our
method is the plant static modeling of the olive tree
from multi-view stereo reconstruction method and
terrestrial LiDAR scanning. Both geometry models
may be the input for our framework which is based on
multispectral image projection from multiple views.
Data from narrow-band views are used to carry out an
early segmentation of the point cloud, in order to sim-
plify the number of features used in the reconstruc-
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
362

tion. In the following, an inverse procedural modeling
is performed on the point cloud to overcome recon-
structing issues as self-hidden branches and leaves of
the plant. To this end, for each view a reflectance
map is computed for the automatic model segmen-
tation. This technique is useful to determine several
constraints for the guided structure generation. We in-
tegrate this into a rule-based growing system, that also
uses a variation of space colonization (Runions et al.,
2007) and shadow-propagation (Palubicki, 2012). As
our method is based on the change in light distribution
from several spectral bands, the illumination of differ-
ent part of the tree is calculated and used to improve
the data-driven reconstruction. In addition, the spatio-
temporal analysis of the plant behavior provides us
real historic data for the growth prediction.
4 POINT CLOUD
RECONSTRUCTION
One of the main steps of our solution consists of
obtaining three-dimensional information from real
plants to complete it with data obtained from mul-
tispectral sensors. For this purpose, several known
techniques might be applied to generate 3D models
of existing real-world plants. This process can be
carried out for each plant or using batches, depend-
ing on the technology used for capturing the data.
We mainly used two methods: Light Detection and
Ranging (LiDAR), and the image-based reconstruc-
tion. These methods produce distinct results, that is,
point clouds with different attributes and spatial distri-
bution. However, both of them offer spatial data with
enough precision to be used in the following steps of
the modeling.
LiDAR. This 3D scanning technique determines the
distance to a point in space by timing the round trip
time of a light pulse typically fired by a laser diode.
The time that passes until the reflected light is cap-
tured by a detector is timed. Visible (green) or in-
visible (near infrared or NIR) lasers are usually used.
Typical time-of-flight laser scanners can measure the
distance of several hundred points per second (high-
density clouds). Airborne LiDAR can scan a wide
area of terrain in a short period of time, and this is
the main reason for using it with the aim of monitor-
ing the temporal evolution of entire plantations. The
main drawback is that 3D scanning is a process prone
to occlusion, especially when capturing plants (Figure
2). However, scanned plants have a higher resolution
than image-based models, hence, more accurate guide
is generated for the inverse modeling.
In order to obtain the skeleton of a tree from the
Figure 2: Point cloud from terrestrial LiDAR scanner.
scanned point cloud, we follow a similar approach as
the first step of the algorithm presented in (Xu et al.,
2007). Due to the typical incompleteness of the point
clouds produced by LiDAR, additional branches have
to be synthesized to complete the tree topology, es-
pecially the crown. We use cluster edges in a span-
ning graph to reconstruct the tree skeleton. However,
leaves are not randomly added to the fine branches as
presented in (Xu et al., 2007). Instead, we rely only
on the scanned data, because our method is targeted
to the monitoring of actual leaves. Our experiments
were mainly targeted to olive trees. This type of plant
is formed by many branches and a very dense crown
with multitude of leaves of different sizes. This ag-
gravates the occlusion of the inner part of the tree.
Multi-view Stereo Reconstruction. This method
supposes a simplification of the technique of stereo
vision for the case of a single camera that takes im-
ages from different positions. Computer vision tech-
niques are used to calculate matching points, since
the positions of the camera do not have scale and
orientation information in relation to the object to
be scanned. This low cost technique is based on
an adjustment procedure that uses a database of fea-
tures. Those features are automatically extracted from
a set of multiple overlapping images. We have used
a combination of SfM and Patch-based MultiView
Stereo (PMVS) to obtain dense point clouds from
trees. The data obtained with this method can com-
plete the data obtained with LiDAR when necessary,
especially with occluded zones at low heights. SfM is
a technique for producing dense point clouds based on
a feature matching process. However, it does not pro-
duce accurate results on plant images with repetitive
or similar feature properties (Guo et al., 2018). For
this reason, we only use the part of SfM that estimates
the stereo camera position for each of the shots. Then,
PMVS produces a point cloud based on the features
found, taking into account the visibility constraints.
This combination of SfM and PMVS produces better
results, but it still has problems with occlusions and
the performance does not scale well. In order to im-
prove the process, a novelty of our method consists
of using images from the multispectral camera for an
Accurate Plant Modeling based on the Real Light Incidence
363

early segmentation of points, based on multispectral
analysis. This step allows us to extract only the points
corresponding to trunks and branches, omitting all
the leaves, which are precisely the part of the plant
that causes all the feature matching issues. After the
views matching, all points are used for the resulting
dense point cloud (Figure 3). This process greatly
increases the precision of the multi-view reconstruc-
tion, especially with overlapped trees. Later, we use
a point clustering process for the inverted modeling
of the tree skeleton. We also plan to incorporate the
method presented in (Guo et al., 2018). It uses a depth
map based reconstruction algorithm. For each view,
a dense depth map is computed. Then, all maps are
projected into a single point cloud model by consid-
ering visibility. Our contribution at this point involves
the addition of multispectral information to the depth
maps. It provides the removal of all superfluous infor-
mation from each map, increasing the performance of
the process.
Figure 3: Point cloud from Structure-from-Motion.
5 PLANT-LIGHT INTERACTION
The surrounding environment plays a key role for
the growing process of trees. The crown shape and
branching structure are directly determined by several
environmental effects. The light energy is considered
as the main factor for branch shooting and it is calcu-
lated by sampling the environmental space. For this
purpose, plant models are decomposed into a grid of
voxels. In this way, a coarse estimation of the inter-
action of light with each bud is computed (Palubicki
et al., 2009). In general, the exposure of each bud to
light is an estimation based on the plant space classi-
fication into a grid of voxels. In this way, the shadow
propagation method is applied to calculate the bud
fate by selecting the adjacent voxel with the lowest
shadow value. In terms of realistic plant behaviour
according to the light sensitivity, multispectral sen-
sors may be used to capture the reflected energy in
various spectral bands.
In this work we have targeted the olive tree as the
main objective. It has been observed by four spectral
bands: Green (530nm-570nm), NIR (770nm-810nm),
Figure 4: Narrow-band channels for each capture.
Red (640nm-680nm) and REG (730nm-840nm) (Fig-
ure 4). For each one, the influence of the sunlight
is studied for multiple purposes: (1) the 3D recon-
struction of the tree structure from multi-views, (2)
the plant space decomposition and (3) the prediction
of natural growth process. To this end, our method
uses as input either some geometric tree models by
LiDAR scans or multiple overlapping images. Once
the 3D point cloud has been extracted, reflectance
maps are computed for each band. Unlike previous
works, which are based on a coarse light propagation,
we calculate the actual light reflectance value (LRV)
for each plant from multispectral image processing.
Following, a backward projection is carried out for
point cloud labeling (Figure 5). For this purpose, the
fisheye distortion model of narrow-band images is ap-
plied for mapping the distorted pixel coordinates to
the 3D points.
Figure 5: The multispectral images are back-projected to
the input point-cloud.
Plant Segmentation. Since the input point cloud may
not be completed and many branches have not been
reconstructed due to the occlusion with each others,
a novel 3D classification method is proposed. It is
based on the inclusion of reflectance values to com-
plete the branching structure and the foliage. As
plants become healthier, the intensity of reflectance
increases in the NIR band and decreases in the Red
band, which is the physical basis for most vegetation
indices. NDVI (Normalized Difference Vegetation
Index) value arguably indicates the plant vigor and
is also effective for distinguishing vegetation from
branches, trunk and soil. As Figure 6 shows, the olive
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
364

tree is subdivided in two classes which correspond
to the foliage and the ramification of branches. This
method provides the capability to reduce the noise of
the 3D model which is produced by the reconstruction
of leaves and narrow branches. Therefore, several im-
provements are in progress aimed for plant modeling
from multi-view images by considering the search of
key points specifically in branches and trunk.
Figure 6: Plant model segmentation.
Plant Space Decomposition. In addition, instead of
performing a voxelization of the plant space, we carry
out a semantic subdivision of the model for studying
shadow propagation. According to previous plant dis-
semination, a 3D clustering method is proposed for
the inverse modeling of the tree structure (Figure 7).
We focus on the classification of several point groups
to identify the plant skeleton and improve the guided
modeling of the real tree shape. In the image below,
the soil, trunk and main branches are joined to the
same cluster. This group shares similar reflectance
values, ranging from 0 to 0.4 and several soft geo-
metric constraints such as distance, normal direction,
cluster volume, etc. Moreover, the plant crown is sub-
divided into six clusters to estimate the canopy den-
sity and the spanning for each branch.
Figure 7: Semantic classification of the plant shape.
Procedural Modeling. The influence of the sur-
rounding environment should be considered for the
prediction of the plant growth. In this context, the pro-
cedural modeling is capable to simulate the adaptivity
of trees by considering several environmental effects.
In general, the space colonization and the shadow
propagation are the most used methods. However, un-
like previous works, which are based on a coarse es-
timate of bud exposure to calculate the optimal grow-
ing direction, our method takes the real light inci-
dence and the photosynthetic activity of plants from
multispectral imagery. Consequently, it provides sev-
eral parameters to predict the next growth stage ac-
cording to the reflectance index which may determine
the pruning factor, phototropism and gravitropism,
branching angle, etc. Our hypothesis is based on es-
timating the growth rate for each region of the plant
due to some parts grow faster than others.
6 CONCLUSIONS AND FUTURE
WORK
We have introduced main contributions of our re-
search in progress. A novel framework is provided
for plant modeling according to the plant-light inter-
action. The key feature of our method is the com-
bination of geometry plant models to semantic data
from multispectral imagery. In summary, plant mod-
eling without the influence of the surrounding envi-
ronment results to static and unrealistic tree repre-
sentations. This work is focused on the olive tree
structure and their interactions with the environmen-
tal light. In this paper, several techniques for plant
modeling and novel methods based on spatial clas-
sification have been applied to estimate the growing
shoot from the light-based plant behaviour. To this
end, a plant segmentation is carried out to dissemi-
nate the branching structure to the foliage. Then, a
semantic and volumetric plant space decomposition
is proposed for spatial pooling by several features in
common. Moreover, a historic set of spectral images
have been acquired to predict next growing stages of
target trees from the reflectance absorption.
Several open problems may be approached for fu-
ture work. For the monitoring of extensive planta-
tions, it is not feasible to store a dense point cloud
for every plant or bush. First, we would general-
ize our method to model larger scale scenes, such as
real ecosystem of even a forest. Second, since the
plant space decomposition, we focus on generating a
rule-based system to reconstruct self-hidden geome-
try from previous knowledge of how each plant re-
gions interact with the environment. Finally, the de-
velopment of a robust predictive model based on mul-
tispectral or hiperspectral data to simulate the grow-
ing process under different environmental effects.
Accurate Plant Modeling based on the Real Light Incidence
365

ACKNOWLEDGEMENTS
This work has been partially supported by the Min-
isterio de Econom
´
ıa y Competitividad and the Euro-
pean Union (via ERDF funds) through the research
project (TIN2017-84968-R).
REFERENCES
Bene
ˇ
s, B., Stava, O., M
ˇ
ech, R., and Miller, G. (2011).
Guided procedural modeling. In Computer graphics
forum, volume 30, pages 325–334. Wiley Online Li-
brary.
Berger, M., Tagliasacchi, A., Seversky, L. M., Alliez, P.,
Guennebaud, G., Levine, J. A., Sharf, A., and Silva,
C. T. (2017). A survey of surface reconstruction from
point clouds. In Computer Graphics Forum, vol-
ume 36, pages 301–329. Wiley Online Library.
Candiago, S., Remondino, F., De Giglio, M., Dubbini,
M., and Gattelli, M. (2015). Evaluating multispec-
tral images and vegetation indices for precision farm-
ing applications from uav images. Remote Sensing,
7(4):4026–4047.
Deussen, O., Hanrahan, P., Lintermann, B., M
ˇ
ech, R., Pharr,
M., and Prusinkiewicz, P. (1998). Realistic modeling
and rendering of plant ecosystems. In Proceedings of
the 25th Annual Conference on Computer Graphics
and Interactive Techniques, SIGGRAPH ’98, pages
275–286, New York, NY, USA. ACM.
Guo, J., Xu, S., Yan, D.-M., Cheng, Z., Jaeger, M., and
Zhang, X. (2018). Realistic procedural plant model-
ing from multiple view images. IEEE transactions on
visualization and computer graphics.
Hakkenberg, C., Peet, R., Urban, D., and Song, C. (2018).
Modeling plant composition as community continua
in a forest landscape with lidar and hyperspectral re-
mote sensing. Ecological Applications, 28(1):177–
190.
Klodt, M. and Cremers, D. (2014). High-resolution plant
shape measurements from multi-view stereo recon-
struction. In European Conference on Computer Vi-
sion, pages 174–184. Springer.
Livny, Y., Yan, F., Olson, M., Chen, B., Zhang, H., and
El-Sana, J. (2010). Automatic reconstruction of tree
skeletal structures from point clouds. ACM Trans.
Graph., 29(6):151:1–151:8.
Lou, L., Liu, Y., Han, J., and Doonan, J. H. (2014). Ac-
curate multi-view stereo 3d reconstruction for cost-
effective plant phenotyping. pages 349–356.
Lowe, D. G. (2004). Distinctive image features from scale-
invariant keypoints. International journal of computer
vision, 60(2):91–110.
Moran, M. S., Inoue, Y., and Barnes, E. (1997). Opportuni-
ties and limitations for image-based remote sensing in
precision crop management. Remote sensing of Envi-
ronment, 61(3):319–346.
Omasa, K., Hosoi, F., and Konishi, A. (2006). 3d li-
dar imaging for detecting and understanding plant re-
sponses and canopy structure. Journal of experimental
botany, 58(4):881–898.
Oppenheimer, P. E. (1986). Real time design and anima-
tion of fractal plants and trees. In ACM SiGGRAPH
Computer Graphics, volume 20, pages 55–64. ACM.
Palubicki, W. (2012). Fuzzy Plant Modeling with OpenGL:
Novel Approaches in Simulating Phototropism and
Environmental Conditions. AV Akademikerverlag.
Palubicki, W., Horel, K., Longay, S., Runions, A., Lane,
B., M
ˇ
ech, R., and Prusinkiewicz, P. (2009). Self-
organizing tree models for image synthesis. In ACM
Transactions on Graphics (TOG), volume 28, page 58.
ACM.
Pe
˜
nuelas, J. and Filella, I. (1998). Visible and near-infrared
reflectance techniques for diagnosing plant physiolog-
ical status. Trends in plant science, 3(4):151–156.
Prusinkiewicz, P. (1986). Graphical applications of l-
systems. In Proceedings of graphics interface, vol-
ume 86, pages 247–253.
Prusinkiewicz, P. (2004). Modeling plant growth and devel-
opment. Current Opinion in Plant Biology, 7(1):79 –
83.
Quan, L., Tan, P., Zeng, G., Yuan, L., Wang, J., and Kang,
S. B. (2006). Image-based plant modeling. In ACM
Transactions on Graphics (TOG), volume 25, pages
599–604. ACM.
Runions, A., Lane, B., and Prusinkiewicz, P. (2007). Mod-
eling trees with a space colonization algorithm. NPH,
7:63–70.
Sankaran, S., Mishra, A., Ehsani, R., and Davis, C. (2010).
A review of advanced techniques for detecting plant
diseases. Computers and Electronics in Agriculture,
72(1):1–13.
Stava, O., Benes, B., Mech, R., Aliaga, D. G., and Kristof,
P. (2010). Inverse procedural modeling by automatic
generation of l-systems. Comput. Graph. Forum,
29(2):665–674.
Stava, O., Pirk, S., Kratt, J., Chen, B., M
ˇ
ech, R., Deussen,
O., and Benes, B. (2014). Inverse procedural mod-
elling of trees. In Computer Graphics Forum, vol-
ume 33, pages 118–131. Wiley Online Library.
Suh, H. K., Hofstee, J. W., and van Henten, E. J. (2018). Im-
proved vegetation segmentation with ground shadow
removal using an hdr camera. Precision Agriculture,
19(2):218–237.
Thomas, S., Kuska, M. T., Bohnenkamp, D., Brugger,
A., Alisaac, E., Wahabzada, M., Behmann, J., and
Mahlein, A.-K. (2018). Benefits of hyperspectral
imaging for plant disease detection and plant protec-
tion: a technical perspective. Journal of Plant Dis-
eases and Protection, 125(1):5–20.
Xu, H., Gossett, N., and Chen, B. (2007). Knowledge and
heuristic-based modeling of laser-scanned trees. ACM
Trans. Graph., 26(4).
Yi, L., Li, H., Guo, J., Deussen, O., and Zhang, X. (2015).
Light-Guided Tree Modeling of Diverse Biomorphs.
In Stam, J., Mitra, N. J., and Xu, K., editors, Pa-
cific Graphics Short Papers. The Eurographics Asso-
ciation.
GRAPP 2019 - 14th International Conference on Computer Graphics Theory and Applications
366
