
Unsupervised Image Segmentation using Convolutional Neural Networks
for Automated Crop Monitoring
Prakruti Bhatt, Sanat Sarangi and Srinivasu Pappula
TCS Research and Innovation, Mumbai, India
Keywords:
Unsupervised Segmentation, Severity Measurement, Crop Monitoring.
Abstract:
Among endeavors towards automation in agriculture, localization and segmentation of various events during
the growth cycle of a crop is critical and can be challenging in a dense foliage. Convolutional Neural Network
based methods have been used to achieve state-of-the-art results in supervised image segmentation. In this
paper, we investigate the unsupervised method of segmentation for monitoring crop growth and health condi-
tions. Individual segments are then evaluated for their size, color, and texture in order to measure the possible
change in the crop like emergence of a flower, fruit, deficiency, disease or pest. Supervised methods require
ground truth labels of the segments in a large number of the images for training a neural network which can
be used for similar kind of images on which the network is trained. Instead, we use information of spatial
continuity in pixels and boundaries in a given image to update the feature representation and label assign-
ment to every pixel using a fully convolutional network. Given that manual labeling of crop images is time
consuming but quantifying an event occurrence in the farm is of utmost importance, our proposed approach
achieves promising results on images of crops captured in different conditions. We obtained 94% accuracy in
segmenting Cabbage with Black Moth pest, 81% in getting segments affected by Helopeltis pest on Tea leaves
and 92% in spotting fruits on a Citrus tree where accuracy is defined in terms of intersection over union of the
resulting segments with the ground truth. The resulting segments have been used for temporal crop monitoring
and severity measurement in case of disease or pest manifestations.
1 INTRODUCTION
Robotic farm surveillance, automatic process control,
and automated advisory for any event in the farms are
becoming extremely important to increase quality of
food production all over the world given the incre-
asing population and limited availability of resour-
ces such as agricultural experts and farm labor. To
achieve this, automated plant phenotyping is impor-
tant, and precise segmentation is a key task for this.
Presence of occlusions, variability in shapes, shades,
angle and imaging conditions like background and
lighting make it challenging. In most of the crops,
generally the diseases and deficiencies manifest them-
selves as yellowing or browning of leaves in form of
patches or spots and scorching. Also, different parts
of the plant usually have different shapes, textures or
colors. Presence of insects and pests can also be vi-
sibly identified. Image processing techniques have
been used to detect color and texture of the disease
affected area (Singh and Misra, 2017), (Wang et al.,
2008) on a single leaf placed at the center of a plain
background.
Considering that the deep convolutional networks
based approaches have surpassed other approaches in
terms of accuracy, various CNN based methods have
been applied in leaf segmentation (Aich and Stavness,
2017), (He et al., 2016), (Pound et al., 2017) with im-
pressive results in segmenting out the leaves. These
methods are fully supervised where the CNN based
models are trained on annotated data of images cap-
tured with similar resolution, same lighting and back-
ground conditions in the lab. In a real scenario howe-
ver, the images would be with different backgrounds,
occlusions, overlap and lighting conditions. Parts of
plants have reasonable amount of variation especially
in different growth stages. Applying supervised met-
hods for segmentation of multiple crops in different
regions would need a huge amount of annotated data.
Variations in the crop at different stages and multiple
manifestations of a disease or pests in different sizes
make the annotations and data collection a time con-
suming task. Hence, we propose to use unsupervised
methods to segment the images in order to make it
applicable to multiple crops and use-cases. We pro-
pose a novel system comprising (i) an unsupervised
Bhatt, P., Sarangi, S. and Pappula, S.
Unsupervised Image Segmentation using Convolutional Neural Networks for Automated Crop Monitoring.
DOI: 10.5220/0007687508870893
In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2019), pages 887-893
ISBN: 978-989-758-351-3
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
887

method for segmentation based on a fully convolutio-
nal network as a feature extractor with back propaga-
tion of the pixel labels modified according to the out-
put of a graph based method – Normalized cut (Shi
and Malik, 2000), and (ii) a method to identify dif-
ferent segments based on color, texture, and size to
closely monitor different plants in indoor and outdoor
farms. We have presented the results for segmenta-
tion of parts of plants, and disease and pest affected
regions in them for 6 different crops viz. yellowing in
Variagated Balfour Aralia and Dracaena, Helopeltis
pest in Tea leaves, Black Moth in Cabbage, Anthra-
cnose in Pomegranate, and fruit in a Citrus tree.
2 RELATED WORK
Supervised image segmentation methods (Farabet
et al., 2013), (Badrinarayanan et al., 2017), (Ron-
neberger et al., 2015), (Hariharan et al., 2014) based
on CNN have been widely used for many applications
like autonomous vehicles and medical image analysis.
These methods have achieved state-of-the-art results
in semantic as well as instance level segmentation, but
these models require to be trained with a large number
of images along with their ground truth annotations at
the segment level. Weakly supervised methods have
also been proposed where the training data for seman-
tic segmentation is a mixture of a few object segments
and a large number of bounding boxes (Chang et al.,
2014), or the dataset only contains the class speci-
fic saliancy maps (Shimoda and Yanai, 2016). Re-
cently, unsupervised methods for obtaining segmen-
tation maps have been proposed in (Kanezaki, 2018)
and (Xia and Kulis, 2017). In (Kanezaki, 2018),
the cluster labels of the pixels in a super-pixel obtai-
ned by SLIC are corrected and used for back propoga-
tion to train the convolutional blocks. Authors in (Xia
and Kulis, 2017) have used two U-Nets (Ronneberger
et al., 2015) as an autoencoder, where encoding layer
produces a pixelwise prediction and post-processing
involving Conditional Random fields (CRF) and hier-
archical segmentation for the encoder end to segment
the image.
Fully convolutional networks (FCNs) (Long et al.,
2015) have been proven as effective for solving the
semantic segmentation problem. One advantage of
using them is that images of arbitrary size can be input
to the network and the segmentation map of the same
size can be obtained. Conditional Random fields have
been applied to smoothen the segmented boundaries.
Liu et. al in (Liu et al., 2015) have used CRF as a
post-processing step after the inference from CNN to
refine the segmentation map. Chen et al. in (Chen
et al., 2015) have proposed to train a FCN followed
by fully connected gaussian CRF to accurately model
the spatial relationships of the pixels in the images.
We perform Unsupervised segmentation using a
FCN, and jointly optimize the image features and
cluster label assignment for any image given as in-
put. The pixel groups obtained through adjacency in-
formation using normalized cut once over the image
is used to update the image features by updating the
network weights.
3 METHOD
The aim is to obtain possible segments from the
image based on pixel features in unsupervised man-
ner. These segments can be further used to make
an understanding out of the image. These features
are different for every image and generally depen-
dent on the color, edges and texture of pixel groups
in the image. Such groups of pixels with similar fe-
atures constitute a segment whose label is unknown
in our case. These features are calculated using the
convolutional network in our application. Consider
{x
n
∈ R
d
}
N
n=1
as a d-dimensional feature of an input
image I with {p
n
∈ R
3
}
N
n=1
pixels and let {l
n
∈ Z}
N
n=1
be the segment label assignment for each pixel where
N is the total number of pixels in the image. The
task of getting this unknown number of labels for
every pixel can be formulated as l
n
= f (x
n
) where
f : R
d
→ Z is the cluster assignment function. For
a fixed x
n
, f is expected to give the best possible la-
bels l
n
. When we train the neural network to learn x
n
and f for a fixed and known set of labels l
n
, it can be
termed as supervised classification. However, in this
paper, we aim to predict the unknown segmentation
map l
n
while iteratively updating the function f and
the features x
n
. Effectively, we jointly
1. predict the optimal l
n
for an updated f and x
n
2. Train the parameters of neural network to get f
and x
n
for the fixed l
n
.
Humans tend to create segments according to the
common salient properties of the objects or patches
in the image like colors, texture, shape. Hence, a seg-
mentation method should also be accurately grouping
spatially continuous pixels having such similar pro-
perties into same class or label. Also, it must assign
different labels to the pixels having different featu-
res. So as in (Unnikrishnan et al., 2007) (Kanezaki,
2018) (Xia and Kulis, 2017), we also apply the fol-
lowing criteria in our method: (i) Pixels with similar
features must be assigned same label. (ii) Spatially
continuous pixels are desired to be having same clus-
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
888
ter label. (iii) Large number of unique labels is desi-
red so that even the smaller segments are not missed.
These criteria obviously have a trade-off and need to
be balanced for good segmentation results. Our way
of jointly optimizing them gives reasonably good re-
sults i.e l
n
for application in images captured for crop
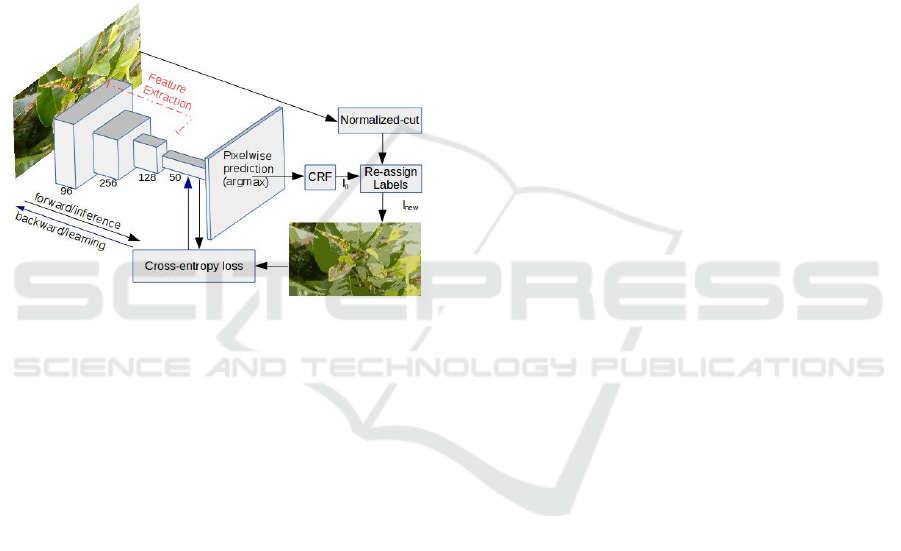
monitoring. Figure 1 illustrates the implemented met-
hod for unsupervised segmentation of crop images.
The size of FCN in the diagram are the ones that were
used in the implementation for the results shown in
this paper. The length of the network and the size of
filters can be changed for better performance. Fol-
lowing subsections and the Algorithm 1 explain the
method in detail.
Figure 1: Block Diagram of the Proposed Approach.
3.1 Feature Calculation and Label
Assignment
We calculate the d dimensional feature vector {x
n
}
N
1
for the pixels of image I using Fully convolutio-
nal network architecture built using locally connected
layers, such as convolution and pooling. The downs-
ampling path in the neural network architecture cap-
tures the semantic information within the image and
the upsampling path helps to recover the spatial infor-
mation (Long et al., 2015). B blocks of convolutio-
nal layers followed by pooling are used to calculate
the features. The output after these convolutional lay-
ers is termed as the features or feature-map {x
n
}
N
n=1
of the image I and can be as {x
n
= W
c
p
n
+ b
c
}
N
n=1
.
After these convolutional blocks, the last convoluti-
onal layer 1 × 1 × M fully convolutional layers are
used for classification of pixels into different clusters,
where M can be considered as the maximum possi-
ble unique labels in which the pixels can be cluste-
red. The response-map of this layer is denoted as
{W
op
x
n
+ b
op
}
N
n=1
. After applying the batch norma-
lization, we obtain the response map {y
n
∈ R
M
}
N
n=1
that has M dimensional vector of values with zero
mean and unit variance for every pixel in the image.
Using this helps to achieve a higher number of clus-
ters, thus satisfying the third criterion as mentioned
in Section 3. The index of the value that is maximum
in y
i
can be considered as the label for the i
th
pixel.
It can be obtained by connecting argmax layer at the
output. The total number of unique segments or the
labels assigned in an image are between 1 to M and
is determined by the image content and training of the
neural network at every iteration. Since the convoluti-
onal networks are known to learn generalized features
in the images well, they help satisfy the first criterion
of assigning same cluster label to same pixels. Post-
processing step of using CRFs on the map of cluster
labels help in increasing the segmentation accuracy
by refining boundaries.
3.2 Label Re-assignment and Feature
Update
For every image, the network self-trains in order to
segment it into certain number of clusters. After ma-
king the inference at every iteration, the labels of
pixels in every superpixel is re-assigned on the pre-
mise that all pixels in a superpixel belong to same
segment. We apply the normalized cut method on
the same image to find the superpixel output. While,
we have used Region Adjacency Graph (RAG) al-
ong with Normalized Cut (Shi and Malik, 2000) on
the image, any super-pixel algorithm (Achanta et al.,
2012), (Felzenszwalb and Huttenlocher, 2004) can be
used to obtain the over-segmented map of an image.
The super-pixel level output of the normalized cut
method applied on the same image is used to up-
date the cluster assignment denoted by {l
new
n
}
N
n=1
of
pixels in every super-pixel. After prediction from
neural netwok, the pixels belonging a particular su-
perpixel might have different cluster labels denoted
by {l
n
}
N
n=1
. All these pixels are re-assigned the single
label possesed by maximum number of pixels in that
superpixel. These updated cluster labels are then used
for backpropogation to train the feature extraction
network. The cross entropy loss is calculated between
the response-map {y
n
∈ R
M
}
N
n=1
and the super-pixel
refined cluster labels {l
n
ew}
N
n=1
and then backpropa-
gated to update the weights of the FCN for say T ite-
rations. Using superpixels helps to compute features
on more meaningful regions. They help to get disjoint
partitions of the image and preserve image bounda-
ries. Also, every superpixel is expected to represent
connected sets of pixels. This helps to satisfy the se-
cond criterion of having spatially continuous pixels
in same segment. Here, training of the neural net-
work involves the learning of parameters {W
c
, b
c
} of
Unsupervised Image Segmentation using Convolutional Neural Networks for Automated Crop Monitoring
889

the convolutional layers of the FCN that contributes
in getting image features and also {W
op
, b
op
}, the pa-
rameters of output 1 × 1 ×M layer used to get cluster
map for each pixel. This is termed as feature updation
as shown in the step 10 of the Algorithm 1
Algorithm 1: Unsupervised Segmentation.
Input: I = {p
n
}
N
n=1
in R
3
Output: Labels {l
n
}
N
n=1
1: {S
k
}
K
k=1
← NormCut({p
n
}
N
n=1
)
2: for i from 1 to T : do
3: {x
n
}
N
n=1
← GetFeatures(I, {W
c
, b
c
})
4: {y
n
}
N
n=1
← BatchNorm(W
op
∗ x
n
+ b
op
)
5: {l
n
}
N
n=1
← argmax({y
n
}
N
n=1
)
6: {l
n
}
N
n=1
← CRF({l
n
}
N
n=1
, {p
n
}
N
n=1
)
7: for i from 1 to K: do
8: l
new
n
← argmax|l
n
|∀n ∈ S
k
9: loss ← CrossEntropy({y
n
, l
new
n
}
N
n=1
)
10: {W
c
, b
c
, W
op
, b
op
} ← UpdateFeatures(loss)
4 RESULTS
In accordance to the block diagram in Figure 1, the
considered neural network for experiments has 4 con-
volutional layers. More layers of different filter di-
mensions, pooling and upsampling can be used to eva-
luate if they give better feature learning. The last con-
volutional layer is 1×1×50 where 50 can be assumed
as maximum possible number of labels. CRF is used
as the post processing step for refining the segments.
The over-segmented map for the final step of refi-
ned cluster assignment is obtained by using Norma-
lized Cut over the region adjacency graph (RAG) of
the image. Cross-entropy loss between the predicted
output of the last convolutional layer and the refined
labels is calculated for backpropagation. Stochastic
Gradient Dscent with a learning rate of 0.01 and mo-
mentum of 0.9 is used to train the neural network for
T = 500 iterations. We have evaluated the segmen-
tation performance using the measure in Equation 1.
Here, A denotes the accuracy of predicted cluster la-
bels l
n
for all pixels with respect to the ground truth
labels {gt
n
}
N
n=1
, and is defined by ratio of correctly
predicted pixels to the total number of pixels N in the
image.
A(l
n
, gt
n
) =
Number of correct l
n
N
∀n ∈ [1, N] (1)
For images of different crops, we applied the propo-
sed segmentation method and have obtained accepta-
ble results in terms of segments and their count. Co-
lumn (b) in Figure 2 gives the crop-wise segmentation
results using our method where the optimal number
of clusters and segments are obtained. The color of
every segment in the plotted result is the average of
the RGB values of pixels assigned that particular seg-
ment label.
Refering to the images in Figure. 2, we obtained
4 segments (unique labels or clusters) in Variegated
Balfour Aralia with 92% accuracy, 3 segments in Dra-
caena, 7 segments in Tea with 81% accuracy, 6 seg-
ments in Cabbage with 94% accuracy, 9 segments in
Pomegranate with 67% accuracy, 6 segments in Ci-
trus with 93% accuracy. Considering the trade-off be-
tween the number of segments and the way pixels are
assigned the cluster labels, more number of segments
are helpful for detecting any change in an image for
crop monitoring. We could successfully obtain the
segments of interest in all of these images, i.e. yel-
low segments in Aralia and Dracaena, black region in
cabbage due to Black Moth, brown regions in Tea and
Pomegranate due to attack of Helopeltis and Anthra-
cnose respectively, as well as appearance of a fruit in
a Citrus tree.
The main disadvantages in using the conventional
methods of clustering are (a) finding the correct fe-
atures that help in getting correct clusters and (b) the
need to specify the desired number of clusters as input
along with the image. These methods are also sensi-
tive to the imaging conditions like light exposure and
clarity. Column (c) in Figure. 2 shows the segments
obtained by using a conventional method of color ba-
sed clustering (K-means) over the same images. The
number of clusters is fixed to 5 and the images were
converted to CIELAB format before clustering. We
obtained best results collectively for all the conside-
red images when converted to CIELAB format with
the number of clusters fixed to 5 in K-means method.
Even if the image got segmented according to the dif-
ference in pixel values, we did not achieve the seg-
ments of interest. The color of each segment shown
in the results is representative of the average of RGB
values of all pixels in that cluster. The cluster labels
in column (c) of Figure 2 are mixed up and we could
not make out which segment corresponds to the co-
lor change, while the segments of yellow color on the
leaves of Aralia and Dracaena are easily visible in co-
lumn (b) images. Hence the proposed method helps
to achieve better results in plant images by accura-
tely and efficiently assigning the segment labels as
expected.
Once the image is satisfactorily segmented, dif-
ferent ensembles of computer vision methods can be
applied to achieve maximal automation in plant mo-
nitoring as described in the next section.
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
890

1. Aralia
2. Dracaena
3. Tea 4. Cabbage
(a) Plant image
5. Pomegranate
(b) Our method (c) Clustering (a) Plant image
6. Citrus fruit
(b) Our method (c) Clustering
Figure 2: Segmentation results on different plant images with our method and K-means clustering in columns (a), (b) and (c).
5 APPLICATIONS
Our proposed unsupervised segmentation method
while developed specifically for plant images is gene-
ral enough to be applicable in other domains. As dis-
cussed in Sec. 1, the segmentation and measurement
of any event in case of plants is challenging due to
high inter-class variations. Moreover, annotating the
small segments like Helopeltis in tea or Anthracnose
in pomegranate as seen in Figure. 2 is a tedious task.
As mentioned before, most manifestations of diseases
and pests can be identified as change in color and tex-
ture on the crop parts. The real-time performance of
this method enables easy deployments on edge devi-
ces like mobile phones, or camera installations in the
fields.
5.1 Detecting Change in Plant
Appearance
In applications like monitoring plants through a tem-
poral sequence of images, it helps to use change de-
tection methods to identify changes of interest. By
detecting the growth stages of the plant, suggestions
on fertilizers or pesticides can be made. This can
also used to notify agri-experts about the events in
the farm. As seen in Figure. 2, yellowing on leaves
has resulted in an additional yellow segment on the
images of Dracaena leaves and Aralia leaves, brown
spots on tea leaves and pomegranate fruit, and brown
holes on cabbage. The colors of these segments is
identified by using the HSV values of the pixels in the
newly emerged segments. If the color value of the ad-
ditional segments is between yellow and brown, it is
usually an indication that the plant suffers from low
soil moisture or Nitrogen deficiency or is affected by
some pest or disease.
5.2 Estimating Severity of Plant Health
Condition
Pests and diseases contribute to some of the largest
losses in crop yield around the globe. Moreover, due
to lack of knowledge, chemicals are applied either at
wrong growth stage or in wrong quantities. The dise-
ases and pests can be easily detected using existing
classification methods, but the idea of severity and
stage is necessary to take actions at the correct point
of time. Measuring the diseased region out of the
image gives an idea of severity and hence the quan-
tity of the pesticide. Once we obtain the color infor-
mation of all the clusters, we measure the severity of
Unsupervised Image Segmentation using Convolutional Neural Networks for Automated Crop Monitoring
891

the plant according to Equation 2. Here, c
i j
denotes
the i
th
cluster that lies inside the boundary of the j
th
segment of interest.
Severity =
∑
Number of pixels ∈ {c
i j
}
nc
i=1
Total pixels ∈ c
j
(2)
For example, in case of Helopeltis pest in tea leaves
(Figure 2(c)), we consider all the group of pixels that
(i) belong to all the nc labels that have pixels of brown
value ({c
i j
}
2
i=1
) as well as (ii) are inside the leaf boun-
dary i.e. surrounded by pixels with green value (c
j
))
as the set of pixels representing pest on the leaves.
Here nc = 2 as we have two different labels of brown
shade that represent pests, and the severity measure
for it is 11.41%
(a) Temporal increase in yellowing
(b) Segmentation using proposed method
Figure 3: Temporal increase of yellowing in leaves of Vari-
agated Balfour Aralia (left to right).
The images can be analyzed for temporal events
using this method for monitoring the rate of change
in the segmented regions in consecutive images. For
example, the three temporal images in Figure. 3 illus-
trate different health conditions of the same plant i.e.
Aralia. The image in first column is that of a healthy
plant, the middle column has some part of leaf yel-
lowed and the last column has whole leaf in yellow
which is a condition on a higher level of severity. The
severity for these health stages in Aralia is 0% , 7.27%
and 14.29% respectively. The lighter to darker shades
of the segment also show the level of yellowing in
the plant. The colors imply the mean of pixel values
in the corresponding segments. All the experiments
were done using Keras (Chollet, 2015) for building
and training the neural network and OpenCV (Brad-
ski, 2000) for image processing.
6 CONCLUSION
We have proposed an innovative method to carry out
semantic segmentation through unsupervised feature
learning with an FCN followed by CRF. This enables
image segmentation into optimal number of clusters
without any prior information or training. Different
architectures of neural network for feature calculation
as well as various ways of backpropogation can be
explored to evaluate the performance on various ima-
ges. Experimental results on different crop images
prove utility of the method for automated crop moni-
toring which is a challenging application given inter-
class variations. Deployment of the model on the
edge is also feasible for flagging crop related chan-
ges as it works real-time on images taken in uncon-
trolled conditions. In future, we would also evalu-
ate for hyper-parameters and other network structures
that could further enhance the quality of segmenta-
tion in the considered and other datasets. We aim to
evaluate other methods like the pixel adjacency, tex-
ture difference, edges and gradient information to re-
assign the cluster labels in order to get more refined
segments.
REFERENCES
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., and
Ssstrunk, S. (2012). Slic superpixels compared to
state-of-the-art superpixel methods. IEEE Transacti-
ons on Pattern Analysis and Machine Intelligence,
34(11):2274–2282.
Aich, S. and Stavness, I. (2017). Leaf counting with deep
convolutional and deconvolutional networks. IEEE
International Conference on Computer Vision Works-
hops (ICCVW), pages 2080–2089.
Badrinarayanan, V., Kendall, A., and Cipolla, R. (2017).
Segnet: A deep convolutional encoder-decoder ar-
chitecture for image segmentation. IEEE Transacti-
ons on Pattern Analysis and Machine Intelligence,
39(12):2481–2495.
Bradski, G. (2000). The OpenCV Library. Dr. Dobb’s Jour-
nal of Software Tools.
Chang, F., Lin, Y., and Hsu, K. (2014). Multiple structured-
instance learning for semantic segmentation with un-
certain training data. In IEEE Conference on Compu-
ter Vision and Pattern Recognition, pages 360–367.
Chen, L., Papandreou, G., Kokkinos, I., Murphy, K., and
Yuille, A. (2015). Semantic image segmentation with
deep convolutional nets and fully connected crfs. In
Proceedings of the International Conference on Lear-
ning Representations.
Chollet, F. (2015). Keras. https://github.com/fchollet/keras.
Farabet, C., Couprie, C., Najman, L., and LeCun, Y.
(2013). Learning hierarchical features for scene la-
ICPRAM 2019 - 8th International Conference on Pattern Recognition Applications and Methods
892

beling. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 35(8):1915–1929.
Felzenszwalb, P. F. and Huttenlocher, D. P. (2004). Effi-
cient graph-based image segmentation. International
Journal of Computer Vision, 59(2):167–181.
Hariharan, B., Arbel
´
aez, P., Girshick, R., and Malik, J.
(2014). Simultaneous detection and segmentation. In
Fleet, D., Pajdla, T., Schiele, B., and Tuytelaars, T.,
editors, Computer Vision – ECCV 2014, pages 297–
312. Springer International Publishing.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resi-
dual learning for image recognition. IEEE Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 770–778.
Kanezaki, A. (2018). Unsupervised image segmentation by
backpropagation. In Proceedings of IEEE Internatio-
nal Conference on Acoustics, Speech, and Signal Pro-
cessing (ICASSP).
Liu, F., Lin, G., and Shen, C. (2015). Crf learning with cnn
features for image segmentation. Pattern Recognition,
48(10):2983–2992. Discriminative Feature Learning
from Big Data for Visual Recognition.
Long, J., Shelhamer, E., and Darrell, T. (2015). Fully con-
volutional networks for semantic segmentation. In
IEEE Conference on Computer Vision and Pattern Re-
cognition (CVPR), pages 3431–3440.
Pound, M. P., Atkinson, J. A., Wells, D. M., Pridmore, T. P.,
and French, A. P. (2017). Deep learning for multitask
plant phenotyping.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing
and Computer-Assisted Intervention, pages 234–241.
Springer International Publishing.
Shi, J. and Malik, J. (2000). Normalized cuts and image
segmentation. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 22(8):888–905.
Shimoda, W. and Yanai, K. (2016). Distinct class-specific
saliency maps for weakly supervised semantic seg-
mentation. In Leibe, B., Matas, J., Sebe, N., and Wel-
ling, M., editors, Computer Vision – ECCV 2016, pa-
ges 218–234. Springer International Publishing.
Singh, V. and Misra, A. (2017). Detection of plant leaf di-
seases using image segmentation and soft computing
techniques. Information Processing in Agriculture,
4(1):41 – 49.
Unnikrishnan, R., Pantofaru, C., and Hebert, M. (2007). To-
ward objective evaluation of image segmentation al-
gorithms. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 29(6):929–944.
Wang, L., Yang, T., and Tian, Y. (2008). Crop disease leaf
image segmentation method based on color features.
In Li, D., editor, Computer And Computing Technolo-
gies In Agriculture, Volume I, pages 713–717, Boston,
MA. Springer US.
Xia, X. and Kulis, B. (2017). W-net: A deep model for fully
unsupervised image segmentation. CoRR.
Unsupervised Image Segmentation using Convolutional Neural Networks for Automated Crop Monitoring
893
