
Construct Semantic Type of “Gene-mutation-disease” Relation by
Computer-aided Curation from Biomedical Literature
Dongsheng Zhao
a
, Fan Tong
b
and Zheheng Luo
c
Information Center, Academy of Military Medical Science, Beijing, China
Keywords: Semantic Type, Text Mining, Curation, Gene, Mutation, Disease.
Abstract: Background: Current semantic type of “gene-mutation-disease” relation lacks fine-grained classification and
corresponding relation signal words, which limits its usage in relation extraction from biomedical literature
using text mining approach. Methods: We propose a computer-aided curation pipeline in which open relation
extraction, signal word clustering, relation type mapping are used to analyze biomedical abstracts for semantic
type of “gene-mutation-disease” construction. Coverage metrics are used to evaluate the defined relation type
while ClinVar is chosen as a target to test our semantic type’s usability and performance on guiding relation
extraction from biomedical literature. Results: We have constructed a 5-layer and 16-category semantic type
of “gene-mutation-disease” relation with a vocabulary list containing 58 commonly used relation signal words.
The vocabulary list has coverage of 95.08% and the semantic type has coverage of 94.12%. From 25 abstracts
linked to 30 ClinVar records, 15 relations are correctly mapped and 8 novel relations are discovered
additionally. Conclusion: The results show that our semantic type can cover the main relations between “gene”,
“mutation” and “disease” and can achieve good performance on guiding relation extraction from biomedical
text even using relatively out-of-date dictionary-based text mining methods.
1 BACKGROUND
With the development of biotechnology and the
promotion of precision medicine research, “gene-
mutation-disease” relations have been broadly
studied recently, resulting in over 10 thousand
published papers each year (Allahyari et al., 2017;
Burger et al., 2014). A fraction of these relations has
been collected in domain knowledge database after
meticulous human curation and iterative revise,
which greatly enhanced our understanding towards
disease etiology and pathology (Salgado et al., 2016).
However, there still exists a large amount of valuable
information scattered in numerous literature far
beyond discovery (Rather, Patel and Khan, 2017).
The lack of suitable semantic types and no useable
vocabulary list of “gene-mutation-disease” relation
for biomedical literature mining are two critical
reasons.
Generally, the methods to define semantic type of
“gene-mutation-disease” relation are consistent with
a
https://orcid.org/0000-0003-2616-8891
b
https://orcid.org/0000-0001-6636-8578
c
https://orcid.org/0000-0003-4516-5901
the approaches to construct domain ontology, where
knowledge-based manual definition and semi-
automatic extraction guided by thesaurus and top-
level ontology are frequently used (Bautista-
Zambrana, 2015; Beheshti and Ejei, 2015;
Fernández-López, Gómez-Pérez and Suárez-
Figueroa, 2013). Recent works include the study of
HM Dingerdissen (2017) and J Piñero (2016), and the
most widely accepted and utilized semantic type is
introduced by ClinVar (Landrum et al., 2013). HM
Dingerdissen classified the relation between “gene”,
“mutation” and “disease” into “benign”, “possibly
damaging” and “probably damaging” during the
process of building BioMuta database which took
probability into consideration. J Piñero designated
“susceptibility”, “causal” and “modifying” as “gene-
mutation-disease” relation type when constructing
DisGeNet database which focused on the strength of
the association. According to definition by ClinVar, 9
in 14 relation types were applicable to describe the
relation among “gene”, “mutation” and “disease”,
Zhao, D., Tong, F. and Luo, Z.
Construct Semantic Type of “Gene-mutation-disease” Relation by Computer-aided Curation from Biomedical Literature.
DOI: 10.5220/0007688101230130
In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), pages 123-130
ISBN: 978-989-758-353-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
123
including “Benign”, “Likely benign”, “Uncertain
significance”, “Likely pathogenic”, “Pathogenic”,
“association”, “risk factor”, “protective” and
“Affects”, which considerably extended binary
semantic type (i.e. associate with and not associate
with).
Although these relation types may seem diverse,
they are still far from easy-to-use for mining “gene-
mutation-disease” relations from literature. As a
component of ontology, the semantic type of “gene-
mutation-disease” relations are supposed to be a tree
structure containing hypernymy and hyponymy
instead of flattening into a single level. For example,
“Pathogenic”, “risk factor”, “protective” and
“Benign” can be regarded as hyponymy of
“association” because they express different strength
of association between “gene”, “mutation” and
“disease”. Beyond that, since no relation signal words
are available, experts have to manually assign the
relation type based on their comprehension. Our
study on ClinVar shows less than 40% records of
(gene, mutation, relation, disease) quadruplets can be
located in a single sentence or paragraph in the
associated literature, and the words of relation type
such as “Benign”, “Likely benign” and “Uncertain
significance” seldom appear. Many relations are
scattered through articles or even in the supplements,
and the extraction of this relations can be quite time-
consuming and effort-intensive.
In this paper, we build a multi-layer and fine-
grained semantic type of “gene-mutation-disease”
relation containing 5 layers and 16 categories using
text mining and human curation. We also provide a
vocabulary list with 58 frequently used signal word
belonging to these semantic types. Evaluation shows
the coverage of vocabulary list and semantic type are
95.08% and 94.12% respectively. To study the
usability and performance of our semantic type in
guiding relation extraction from biomedical
literature, we put forward a test by calculating how
many reported relation in ClinVar can be extracted
using our defined semantic type from the ClinVar
linked literature. 15 in 30 records can be correctly
mapped and 8 extra relations are found just using old-
fashioned dictionary-based relation extraction
method. The results show that our relation signal
words vocabulary list and semantic type are
applicable to guide “gene-mutation-disease” relation
extraction and assist “gene-mutation-disease”
relation database extension.
2 METHODS
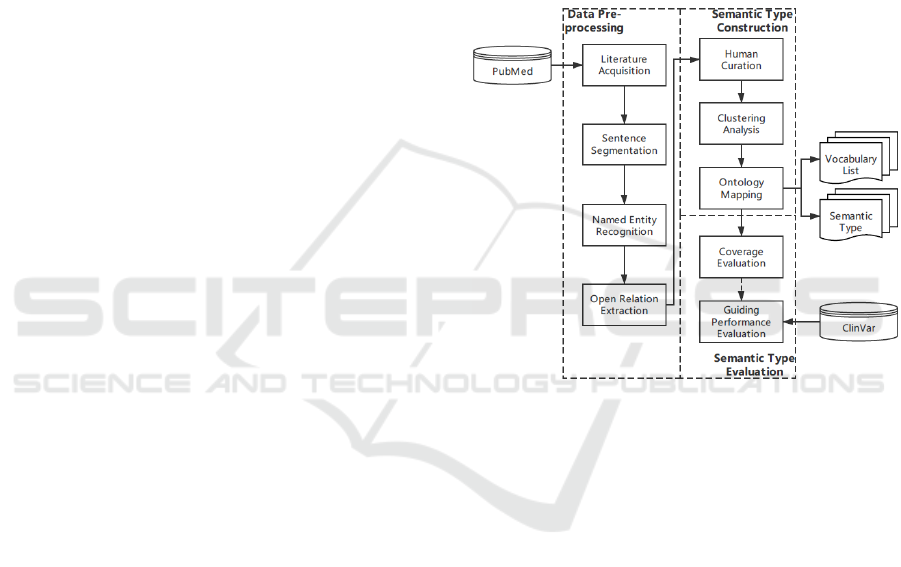
As shown in Figure 1, the pipeline can be divided into
three steps: data pre-processing, semantic type
construction and semantic type evaluation. The first
step mainly deals with the task of data retrieval and
basic natural language processing; the second step
focuses on generating semantic type from the relation
instances found in entities co-occurrence sentences
from the text; the last step evaluates the semantic type
by calculating coverage metrics and testing its
usability and performance on guiding “gene-
mutation-disease” relation extraction.
Figure 1: The overall pipeline of semantic type of “gene-
mutation-disease” relation construction.
2.1 Data Pre-processing
2.1.1 Literature Acquisition
When selecting literature from PubMed as our
preparation dataset, we choose those from the
following three sources: 1) 67 journals with high
impact factor (IF≥ 5.0), 2) PLoS One with large
publication quantity as well as coverage and 3) those
literature correlated to ClinVar databases. We use
“("JournalName"[Journal] AND ("genes"[MeSH
Terms] OR "genes"[All Fields]) AND
("mutation"[MeSH Terms] OR "mutation"[All
Fields]) AND ("disease"[MeSH Terms] OR
"disease"[All Fields]) AND ("2013/01/09"[PDAT] :
"2018/01/07"[PDAT]))” as filtering strategy and
Entrez Programming Utilities as acquisition tools to
get literature for the next step.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
124

2.1.2 Sentence Segmentation
We utilize NLTK (Loper and Bird, 2002) tokenizing
module to split the raw text into sentences as
independent units for named entity recognition and
open relation extraction. Additional syntactic and
statistic rules like the initial letter case in a section and
the length of a section are also applied to correct the
tokenization error from NLTK outputs.
2.1.3 Named Entity Recognition
PubTator (Wei, Kao and Lu, 2013) RESTful API is
invoked for labeling the gene, mutation and disease
mentions. To include more possible related concepts
to make up for the limited number and length of the
obtained abstracts, the expression like “mutation”,
“mutant” and “variant” are also labeled as entities,
which can be used to deal with further co-reference
resolution problem.
2.1.4 Open Relation Extraction
After screening out the “gene-mutation-disease” co-
occurrence sentences, we use Open IE 5.0
(Christensen, Soderland and Etzioni, 2011; Pal, 2016;
Saha and Pal, 2017) to obtain the relation words or
phrases between entities. Unlike domain-specific
relation extraction tools like SemRep (Rindflesch and
Fiszman, 2003), Open IE 5.0 cover a wider range of
potential relations.
2.2 Semantic Type Construction
2.2.1 Human Curation
To revise the results generated by Open IE 5.0, two
experts are asked to curate all the extracted relations
independently according to the following rules: 1) all
modifiers and determiners without critical biomedical
meaning are discarded (“important” in “play an
important role in”); 2) negative expressions are
ignored (“not” in “are not associated with”); 3) the
relation words or phrases are supposed to have simple
present tense. Unanimous choices between the
annotators are chosen to be relation signal words.
2.2.2 Clustering Analysis
Synonyms and homonyms are factors that should be
taken into consideration to minimize the redundancy
of the relation word vocabulary list. The clustering
process is guided by WordNet (Miller and Fellbaum,
2007), a semantic-based vocabulary network. We
first obtain the keyword in candidate relation signal
words. Then, stemming is executed to get the original
form of each keyword, which would then be used to
calculate the similarity between different relation
signal words. We select Leacock and Chodorow score
for evaluation and two signal words having a score
more than 0.5 will be placed into a common word set.
Followed the general-to-specific rule, these scattered
clusters are finally placed on distinctive levels to form
a hierarchical structure.
2.2.3 Ontology Mapping
Due to the limited size of our construction dataset,
even the most commonly-used relation signal words
cannot be ascertained as the best option to describe
the corresponding cluster. By expert consultation, we
find it reasonable to reuse some association relations
from UMLS Semantic Network (McCray, 1989) as
the semantic type of existing cluster, such as “affect”
on the third layer. We choose the top three layers of
UMLS Semantic Network and connect them to more
fine-grained layers derived from clustering analysis,
which finally develops into the semantic type of
“gene-mutation-disease” relation.
2.3 Semantic Type Evaluation
2.3.1 Coverage Evaluation
Coverage is an important metric to assess the
completeness of the domain ontology (Degbelo,
2017). Semantic type, as we mentioned before, is a
component of ontology, which can be evaluated by
coverage test. Respectively, we calculate the
coverage of our defined semantic type by comparing
the results generated from “construction” dataset and
“evaluation” dataset.
2.3.2 Guiding Performance Evaluation
“Distant supervision” takes advantage of related
domain knowledge base as a guidance to make up the
shortage of labeled corpus (Aljamel, Osman and
Acampora, 2015). Inspired by this notion, we put
forward a test which use ClinVar as the target to
calculate how many relations in this knowledge base
can be found using our semantic types. With the
assistance of text mining tools like OpenIE, we obtain
the relation signal word describing the relation of
“gene-mutation-disease” and use the relation signal
words to classify the relation into our defined
semantic types. We build a mapping model from our
semantic type to ClinVar Clinical Significance and
get the overlap between relation defined by us and
curated by ClinVar based on literature linked to
Construct Semantic Type of “Gene-mutation-disease” Relation by Computer-aided Curation from Biomedical Literature
125
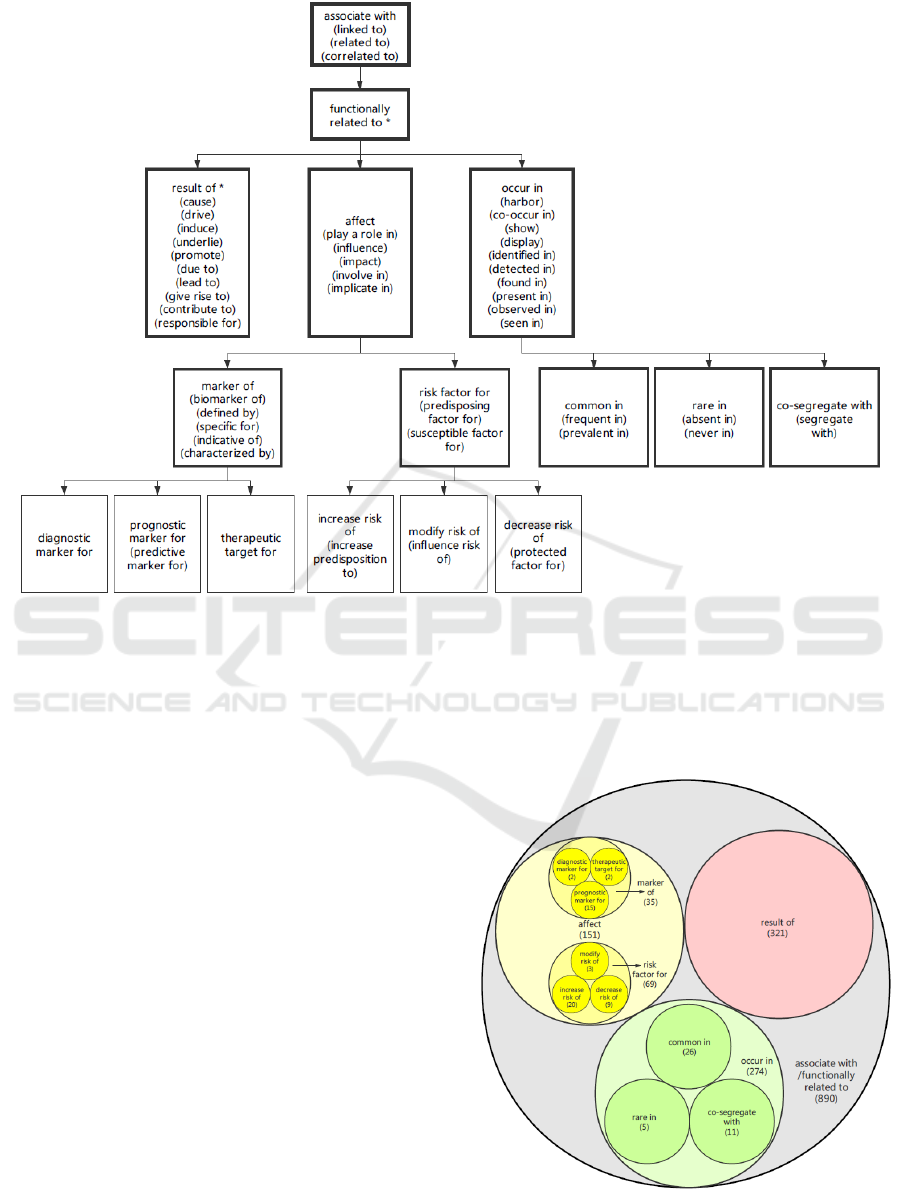
Figure 2: The hierarchy structure and signal word of “gene-mutation-disease” semantic type.
ClinVar in “evaluation” dataset. This result can tell us
how well our semantic type performs in guiding
“gene-mutation-disease” relation extraction.
3 RESULTS
3.1 Semantic Type Construction
After literature acquisition and pre-processing, we got
a total of 570 abstracts in which 336 were from 67
high IF journals, 125 were from PLoS One and 109
were from linked literature to ClinVar. We randomly
selected 513 abstracts as the “construction” dataset
and the remaining 57 literature as “evaluation”
dataset. Through machine processing and human
curation, 890 “gene-mutation-disease” relation
quadruplets were extracted. After filtering, clustering,
and mapping, we eventually constructed a “gene-
mutation-disease” relation semantic type of 5 layers
and 16 categories with 58 commonly used signal
words, as shown in Figure 2. The word in the first line
stood for the semantic type of each set while the
words in bracket refered to its belonging signal
words. For example, in the sentence “The c.626 C >
T (p.P209L) mutation in the BAG3 gene has been
described as causative of a subtype of MFM”,
(BAG3, c.626C > T (p.P209L), cause, MFM) was
extracted which belonged to “result of” semantic
type. The frequency distribution of each semantic
type in curation set is demonstrated in Figure 3.
Figure 3: The frequency distribution of each semantic type.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
126
3.2 Semantic Type Evaluation
3.2.1 Coverage Evaluation
From “evaluation” dataset, we obtained 100 (gene,
mutation, relation, disease) quadruplets. 68 gene
concepts, 35 mutation concepts and 59 disease
concepts were present in these abstracts and each
abstract contained at least one unique relation. The
relatively adequate relations of diverse type extracted
from our samples can be a convincing proof that this
test set was representative enough for a broad range
of biomedical literature.
After manual confirmation, the extracted relation
signal words can be mapped to our defined 58 words
in vocabulary list, with “be etiology of”, “exist in”
and “modifier of” left, which resulted in a coverage
of 95.08% for our vocabulary list. Meanwhile, we
found that “be etiology of” had a similar meaning to
“cause”, while “exist in” was a synonym of “occur
in”. Therefore, 16 relation types other than “modifier”
can be classified into the correct semantic type
automatically or with human effort to extend the
vocabulary list, leading to a 94.12% coverage of our
semantic type. As for “modifier”, we found this word
was similar to “biomarker of” from the morphological
and semantic aspect and could be a subcategory of
“affect”. In this case, this new semantic type could be
added without ruining the overall framework of our
model, which proved the stability and extensibility of
our vocabulary list as well as semantic type.
3.2.2 Guiding Performance Evaluation
Based on the official instruction provided by ClinVar
database, we linked 9 types of ClinVar Clinical
Significance to our model and presented the result in
Table 1. For the category such as “Benign”, “Likely
benign”, “Uncertain significance” and “Likely
pathogenic” which cannot be directly mapped to a
current semantic type, we added negation and
probability description words to existing type for
expressing similar meaning. For instance, from the
sentence “In a six-generation consanguineous
Turkish kindred with both essential tremor and
Parkinson disease, we carried out whole exome
sequencing and pedigree analysis, identifying
HTRA2 p.G399S as the allele likely responsible for
both conditions.”, the obtained relation quadruplet
(HTRA2, p.G399S, responsible for, Parkinson
disease) was classified into “result of” semantic type
in our model. It can be correctly mapped to “Likely
pathogenic” under ClinVar definition due to “likely”,
the modifier word.
For 25 ClinVar linking literature in the
“evaluation” dataset, 30 relations between “gene”,
“mutation” and “disease” were reported by ClinVar
corresponding to the literature. Under the guidance
of our semantic type, we extracted up to 23 (gene,
mutation, relation, disease) quadruplets using our text
mining pipeline. After relation mapping, 15 were
consistent with the records in ClinVar, and 8 novel
were discovered.
The remaining undetected 15 relations can be
attributed to the following reasons: 1) the relation was
not included in abstract section due to its limited
length. For example, in a paper (PMID 25614875),
the relation of (CDKN1C, c.832A>G (p.Lys278Glu),
IMAGe syndrome) was mapped to “Likely
pathogenic” in the abstract while mapped to
“Pathogenic” in the full text; 2) the relation signal
words cannot be located because more than two
mentions were unavailable. For instance, in the
sentence “Three polymorphic variants were identified
in control individuals, of which two were
nonpathogenic (c.1171C > T or p.P391S and c.1413
T > C or p.C471C, with a frequency of 1.5% and 5.5%
respectively) and one pathogenic (c.1330G > C,
frequency 4%).”, although the semantic type of the
relation for the mutation mention, c.1330G > C,
should be “pathogenic” according to the definition of
ClinVar, no signal words can be found according to
our co-occurrence rules.
Table 1: Mapping result from our semantic type to ClinVar
relation type.
Our Model
ClinVar
Examples
Negation + result of
Benign
be
insufficient
to cause
Probability +
Negation + result of
Likely
benign
might not be
a cause of
occur in
Uncertain
significance
co-segregate
with
Probability + result of
Likely
pathogenic
be a probable
driver of
result of
Pathogenic
be
responsible
for
associate with
association
be linked to
risk factor for
risk factor
predispose to
decrease risk of
protective
associate
with reduced
risk of
affect
Affects
be involved
in
8 new pairs of (gene, mutation, relation, disease)
quadruplets we identified were shown in Table 2.
After analysis, we found all these quadruplets came
Construct Semantic Type of “Gene-mutation-disease” Relation by Computer-aided Curation from Biomedical Literature
127

Table 2: Eight (gene, mutation, relation, disease) quadruplets not found in ClinVar.
PMID
Gene
Mutation
Disease
Type of Relation
28487569
ADSL
c.1387-1389delGAG
(p.Glu463Ter)
Adenylosuccinate
lyase deficiency
result of
28487569
ADSL
c.134G>A
(p.Trp45Ter)
Adenylosuccinate
lyase deficiency
result of
26709262
DUOX2
p.A649E
Congenital
Hypothyroidism
result of
26709262
DUOX2
p.R885Q
Congenital
Hypothyroidism
result of
26709262
DUOX2
p.I1080T
Congenital
Hypothyroidism
result of
26709262
DUOX2
p.A1206T
Congenital
Hypothyroidism
result of
26709262
DUOX2
p.Y138X
Congenital
Hypothyroidism
result of
24831256
MYO7A
p.Pro194Hisfs*13
Usher syndrome, type
1
result of
from the literature containing multiple entities and
complex relations, which made it difficult for experts
to locate the relations purely based on the
comprehension. Like in a paper (PMID 28487569),
ClinVar only identified 1 relation between entities but
left 2 potential valuable relations behind. Therefore,
even using a simple dictionary-based relation
extraction method, our model shows great penitential
to assist “gene-mutation-disease” relation knowledge
base construction and extension by automatic
extraction from biomedical literature if a suitable
mapping model is provided. We believe better
performance will be obtained if we use more
advanced text mining methods such as deep learning.
4 DISCUSSION
Our semantic type has been proved to achieve a
relatively good performance, which meets our initial
objective, and can act as a valuable candidate to assist
or guide relation extraction from biomedical
literature. To take a further step, we find following
approaches may contribute to a future improvement
of our model, such as construction dataset
enlargement and mapping model extension.
Originating from relation signal words, our
semantic type relies on the generalization capability
of these words. Take our dataset as an example, when
we only selected the abstracts from PLoS One as
“construction” dataset, “diagnostic marker for” and
“modify risk of” semantic types weren’t generated
due to the absence of corresponding signal type
words. Similarly, the missing semantic type
“modifier” which wasn’t currently included in our
defined semantic type was due to the same reason.
Limited by our strict filtering strategy and number of
experts, we are unlikely to be able to analysis that
large amount of data. But with the increasing scale of
the dataset, we will have access to obtain much more
special relation words. As a result, we believe our
semantic type will achieve better performance.
In addition to lacking fine-grained classification
and corresponding relation signal words, the “gene-
mutation-disease” relation types defined by BioMuta,
DisGeNet and ClinVar are intermediate products
during the process of database construction. Their
relation types are rather isolated and cannot be
directly linked with each other, which makes it
difficult for knowledge integration and sharing. In our
research, the mapping model from ours to ClinVar’s
helped us to locate existing relations and discover
novel relations in the literature. By extending this
mapping model, BioMuta or DisGeNet databases can
provide extra evidence to support these findings.
Take (ADSL, c.1387-1389delGAG (p.Glu463Ter),
result of, Adenylosuccinate lyase deficiency) as an
example, it was extracted but not currently recorded
by ClinVar. By mapping our semantic type “result of”
to DisGeNet “causal” relation type, this relation can
be found and verified in the DisGeNet database.
The future advance of our semantic types can
broaden their usage in the tasks other than biomedical
text mining, such as semantic retrieval from
biomedical knowledge bases. Using proper and
suitable mapping model, our semantic type can bridge
the gap between those isolated knowledge bases and
their linked literature. As a result, a more
comprehensive knowledge base can be developed.
When a relation is searched, our knowledge base can
not only return the related information stored in
ClinVar, BioMuta or DisGeNet, but also provide the
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
128

sentence-level location and context in the linked
literature where the relation originates from. These
detailed results are important for the bioinformatics
researchers who want to grasp an overall
comprehension of their interested entities and
relations.
5 CONCLUSIONS
In this article, focusing on the problem that current
“gene-mutation-disease” semantic types lack fine-
grained classification and corresponding relation
signal words, we propose a text-mining-assisted
semantic type construction approach for automatic
relation extraction from biomedical literature. We
eventually construct a semantic type with 5 layers and
16 categories as well as a corresponding signal word
vocabulary list with 58 commonly-used relation
words. Through coverage and guiding performance
test, even using the old-fashioned dictionary-based
methods, our semantic type is proved not only to have
good performance on coverage evaluation, but also
have great potential in assisting knowledge detection
and discovery from literature. In future works, we
will continue to study deep learning-based solutions
to extract “gene-mutation-disease” relations.
ACKNOWLEDGEMENTS
This Research was funded by National Key R&D
Program of China (2016YFC0901900).
Thanks to Doctor Jiao Li and her team at
Chinese Academy of Medical Sciences for the
guidance in biomedical ontology construction.
REFERENCES
Aljamel, A., Osman, T. and Acampora, G., 2015,
November. Domain-specific relation extraction: Using
distant supervision machine learning. In Knowledge
Discovery, Knowledge Engineering and Knowledge
Management (IC3K), 2015 7th International Joint
Conference on (Vol. 1, pp. 92-103). IEEE.
Allahyari, M., Pouriyeh, S., Assefi, M., Safaei, S., Trippe,
E.D., Gutierrez, J.B. and Kochut, K., 2017. A brief
survey of text mining: Classification, clustering and
extraction techniques. arXiv preprint
arXiv:1707.02919.
Bautista-Zambrana, M.R., 2015. Methodologies to build
ontologies for terminological purposes. Procedia-
Social and Behavioral Sciences, 173, pp.264-269.
Beheshti, M.S.H. and Ejei, F., 2015. Designing and
Implementing Basic Sciences Ontology Based on
Concepts and Relationships of Relevant Thesauri.
Iranian journal of Information Processing &
Management, 30(3), pp.677-696.
Burger, J.D., Doughty, E., Khare, R., Wei, C.H., Mishra,
R., Aberdeen, J., Tresner-Kirsch, D., Wellner, B.,
Kann, M.G., Lu, Z. and Hirschman, L., 2014. Hybrid
curation of gene-mutation relations combining
automated extraction and crowdsourcing. Database,
2014.
Christensen, J., Soderland, S. and Etzioni, O., 2011, June.
An analysis of open information extraction based on
semantic role labeling. In Proceedings of the sixth
international conference on Knowledge capture (pp.
113-120). ACM.
Degbelo, A., 2017, September. A Snapshot of Ontology
Evaluation Criteria and Strategies. In Proceedings of
the 13th International Conference on Semantic Systems
(pp. 1-8). ACM.
Dingerdissen, H.M., Torcivia-Rodriguez, J., Hu, Y., Chang,
T.C., Mazumder, R. and Kahsay, R., 2017. BioMuta
and BioXpress: mutation and expression
knowledgebases for cancer biomarker discovery.
Nucleic acids research, 46(D1), pp.D1128-D1136.
Fernández-López, M., Gómez-Pérez, A. and Suárez-
Figueroa, M.C., 2013. Methodological guidelines for
reusing general ontologies. Data & Knowledge
Engineering, 86, pp.242-275.
Landrum, M.J., Lee, J.M., Riley, G.R., Jang, W.,
Rubinstein, W.S., Church, D.M. and Maglott, D.R.,
2013. ClinVar: public archive of relationships among
sequence variation and human phenotype. Nucleic
acids research, 42(D1), pp.D980-D985.
Loper, E. and Bird, S., 2002, July. NLTK: The natural
language toolkit. In Proceedings of the ACL-02
Workshop on Effective tools and methodologies for
teaching natural language processing and
computational linguistics-Volume 1 (pp. 63-70).
Association for Computational Linguistics.
McCray, A.T., 1989, November. The UMLS Semantic
Network. In Proceedings. Symposium on Computer
Applications in Medical Care (pp. 503-507). American
Medical Informatics Association.
Miller, G.A. and Fellbaum, C., 2007. WordNet then and
now. Language Resources and Evaluation, 41(2),
pp.209-214.
Pal, H., 2016. Demonyms and compound relational nouns
in nominal open ie. In Proceedings of the 5th Workshop
on Automated Knowledge Base Construction (pp. 35-
39).
Piñero, J., Bravo, À., Queralt-Rosinach, N., Gutiérrez-
Sacristán, A., Deu-Pons, J., Centeno, E., Garcia-Garcia,
J., Sanz, F. and Furlong, L.I., 2016. DisGeNET: a
comprehensive platform integrating information on
human disease-associated genes and variants. Nucleic
acids research, p.gkw943.
Rather, N.N., Patel, C.O. and Khan, S.A., 2017. Using deep
learning towards biomedical knowledge discovery. Int.
J. Math. Sci. Comput.(IJMSC), 3(2), pp.1-10.
Construct Semantic Type of “Gene-mutation-disease” Relation by Computer-aided Curation from Biomedical Literature
129

Rindflesch, T.C. and Fiszman, M., 2003. The interaction of
domain knowledge and linguistic structure in natural
language processing: interpreting hypernymic
propositions in biomedical text. Journal of biomedical
informatics, 36(6), pp.462-477.
Saha, S. and Pal, H., 2017. Bootstrapping for Numerical
Open IE. In Proceedings of the 55th Annual Meeting of
the Association for Computational Linguistics (Volume
2: Short Papers) (Vol. 2, pp. 317-323).
Salgado, D., Bellgard, M.I., Desvignes, J.P. and Béroud, C.,
2016. How to identify pathogenic mutations among all
those variations: variant annotation and filtration in the
genome sequencing era. Human mutation, 37(12),
pp.1272-1282.
Wei, C.H., Kao, H.Y. and Lu, Z., 2013. PubTator: a web-
based text mining tool for assisting biocuration. Nucleic
acids research, 41(W1), pp.W518-W522.
BIOINFORMATICS 2019 - 10th International Conference on Bioinformatics Models, Methods and Algorithms
130
