
Study Simulated Epidemics with Deep Learning
Yu-Ju Chen
1
, Tsan-sheng Hsu
2
, Zong-De Jian
2
, Ting-Yu Lin
2
, Mei-Lien Pan
2
and Da-Wei Wang
2
1
Department of CSIE, National Taiwan University, Taipei, Taiwan
2
Institute of Information Science, Academia Sinica, Taipei, Taiwan
Keywords: Agent based Simulation System, Machine Learning, Epidemiology.
Abstract:
Simulation systems are human artifacts to capture the abstraction and simplification of the real world. Study
the output of simulation systems can help us understand the real world better. Deep learning system needs
large volume and high quality data, therefore, a perfect match with simulation systems. We use the data from
an agent based simulation system for disease transmission, to train the deep neural network to perform several
prediction tasks. The model reaches 80 percent accuracy to predict the infectious level of virus, the prediction
of the peak date is off by at most 8 days 90 percent of the time, and the prediction of the peak value is off
at most 20 percent 90 percent of the time at the end of the 7
th
week. We use some preprocessing tricks
and relative error leveling to resolve the magnitude problem. Among all these encouraging results, we did
encounter some difficulty when predicting the index date given information at the middle of an epidemic. We
note that if some interesting concepts are difficult to predict in a simulated world, it sheds some lights on the
difficulty for real world scenarios. To learn the effects of mitigation strategies is an interesting and sensible
next step.
1 INTRODUCTION
The simulation systems can serve as an abstraction
and simplification of real world systems. Many im-
possible to do or hard to do experiments can be carried
out by simulation systems first, so that we can make
more controlled observations cost effectively. The
deep learning technique becomes omnipresent rapidly
in many disciplines, and one of the characteristics is
the monstrous appetite for data, high quality data es-
pecially. Therefore, it is natural to combine simula-
tion systems with deep learning techniques, for exam-
ple there are quite a few interesting results about ap-
plying deep reinforcement learning to gaming(Mnih
et al., 2013).
Agent-based stochastic simulations have been ap-
plied widely for the study of infectious diseases (Ger-
mann et al., 2006). The advantage of the software
simulation models is their flexibility to incorporate
various important concepts in real life compared to
the mathematical models. However, when the pa-
rameter space grows, it becomes more complicated
to draw conclusions from a vast amount of simulated
outcomes. Machine learning models can be seen as
a compression of such vast data, that is, the model
trained is a summary of the data from specific view
point(Li and Vitnyi, 2008).
In this paper, we feed deep learning algorithms
with disease progression data generated by agent-
based simulation systems to study the in silico epi-
demics. One essential question is that if important
characteristics of an epidemic can be estimated or pre-
dicted. For example, the prevalence and the peak date
are important characters of an epidemic(Anderson
and May, 1992). If we can predict them at an early
stage, a better mitigation plan as well as resource al-
location can be developed in time. Above two char-
acteristics are closely related to the infectiousness of
the virus and the stochastic contact patterns of in-
dividuals. In simulation systems, the infectiousness
of the virus is usually modelled by the transmission
probability, denoted p
trans
. The expected behavior of
an epidemic can be estimated by p
trans
and the con-
tacting (mixing) structures. We study the p
trans
pre-
diction problem, which is to predict p
trans
by given
some information about the epidemic. We conduct
a retrospective study to estimate p
trans
after the epi-
demic. The input data for the learning algorithm is
the entire sequence of the number of daily newly in-
fected cases from index date, which is the date first
case appeared. The accuracy of testing is greater than
98%. The model would have higher utility if it can be
applied during the developing phase of an epidemic.
Chen, Y., Hsu, T., Jian, Z., Lin, T., Pan, M. and Wang, D.
Study Simulated Epidemics with Deep Learning.
DOI: 10.5220/0007829702310238
In Proceedings of the 9th International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH 2019), pages 231-238
ISBN: 978-989-758-381-0
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
231

We, therefore, move to the perspective study to pre-
dict the severity of the epidemic at the early stage, 2
to 8 weeks, of an epidemic. We feed first 2 weeks up
to 8 weeks data to train the model, and the accuracy
of the predictions range from around 60% at the end
of 2
nd
week to 78% at the end of 8
th
week.
The next natural question is to estimate the impact
of the endemic, two measurements are widely used in
epidemiology, the peak date and the peak value. The
peak date, the date having the largest number of new
infections, gives a sense of urgency of the epidemic.
The peak date problem is to predict the peak date
given some information of the epidemic. The accu-
racy of testing is around 56% when predicting at the
end of 7
th
week. The predicted peak date is off by at
most 8 days 90% of the time. The other very impor-
tant piece of information for controlling the disease is
peak value, the maximum number of daily newly in-
fected cases in the entire epidemic. The peak value
problem is to predict the peak value. We encounter
two problems; the first is that the peak values range
from a couple 100 up to 600,000 and the second is
that relative error is more appropriate and informative
than absolute error. We use variable length interval to
define our levels. Roughly, the model can predict the
peak value within 10% of error about 70% of times,
and 95% of the time no worst than 20%.
Similar to the weather forecast, we also carry out
the prediction of the number of newly infected cases
for the next day, next day problem. We report the
results to predict the next day when the epidemic still
in early stage (7 to 8 weeks). The accuracy is above
60%, and mean relative error less than 15% and the
prediction is off by less than 10% around 90% of the
time. In real life, there is definitely uncertainty about
the index date, we only can be sure that the index
date is no later than the first observed case. To move
one step closer to cope with the real world scenario,
we study the index date prediction problem, that is
given a sequence of newly infected cases predicting
the index date. Unlike predicting the peak data, this is
a much harder problem, the predicted date has about
40% chance to be in the same week of the true index
date and the prediction is off by at most two weeks
90% of the time.
2 MATERIAL AND METHOD
2.1 Simulation System
We briefly describe how the agent-based simulation
for this study works (Tsai et al., 2010). The core
of the system is a stochastic discrete time agent-
based model. The set of agents, called mock popu-
lation, are people in Taiwan. The mock population,
22.12 millions in size, is constructed according to the
national demographics extracted from Taiwan Cen-
sus 2000 data (http://eng.stat.gov.tw/). Among them,
there are about 1.72 million preschool children (0-5
years old), 2.36 million elementary school children
(6-12 years old), 0.99 million middle school children
(13-15 years old), 0.97 million high school children
(16-18 years old), 3.86 million young adults (19-29
years old), 10.28 million adults (30-64 years old) and
1.94 million elders (65+ years old).
Just like our daily life, two individuals might
have contact because they are family members, co-
workers, classmates, and so on. We use these scenar-
ios to construct our infection mechanism. A proba-
bility is assigned to each pair according to their rela-
tionships, which is captured by contact groups. The
detail of the contact groups will be explained later.
The possibility, effective contact probability, repre-
sents the chance of daily and relatively close contact
which could result in a successful transmission of the
flu virus. An important virus-dependent parameter is
the transmission probability, denoted by p
trans
. It is
the probability that an effective contact results in an
infection. The disease model adopted in the system
is SEIR model. In the model, each individual can
be in one of the following four states, susceptible(S),
exposed(E), infectious(I), and recovered(R). When an
effective contact occurs between an susceptible indi-
vidual and an infectious individual, the susceptible
individual will become exposed with probability of
p
trans
. And according to the disease natural history, an
exposed individual will later become infectious and
then get recovered after antibodies are produced. So
we set the same scenario in our system to make each
individual transform between the four states. In the
simulation system, the average incubation period is
1.9 days and the average infectious period is 4.1 days,
the readers can find detailed information in (Germann
et al., 2006).
A contact group in the setting is a daily close as-
sociation of individuals, where every member is as-
signed an effective contact probability with all other
members in the same group. There are eleven such
contact groups in the model: community, neigh-
borhood, household cluster, household, workgroup,
high school, middle school, elementary school, day-
care center, kindergarten, and playgroup(Chang et al.,
2015). Each individual can belong to several contact
groups simultaneously at any time. The duration of
a simulation run is set as 365 days. Each day has
two 12-hour periods, daytime and nighttime, respec-
tively. During daytime, contact occurs in all contact
SIMULTECH 2019 - 9th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
232
group. School aged children go to schools. There are
around 7.8% school aged children do not go to school
in Taiwan. Preschool children go to a daycare center,
kindergarten, or playgroup. Young adults and adults
are gathered as work-groups. In the nighttime, just
like our daily life that people usually go home after
school or work, contact occurs only in communities,
neighborhoods, household clusters, and households.
The model parameters are similar to ones in a
study by (Germann et al., 2006), with modifications
according to the outcome of a contact diary study in
Taiwan (Fu et al., 2012).
2.2 Data
The epidemic progression data is generated by the
simulation system using the following scenario: each
simulation begins with only selecting 10 random per-
sons from the entire mock population as initial infec-
tious cases at the index date. The only adjustable pa-
rameter is p
trans
here. The range of the p
trans
value
is from 0.075 to 0.105 for all experiments. We di-
vide each level of value by 0.005 and got seven levels
of p
trans
in total. Each level has about 7,000 records,
roughly 50,000 records in total. The training set con-
sists 80% of the records and testing set 20%.
We only use the number of new symptomatic
cases each day for this preliminary exploration. We
do several preprocessing works to tailor the input data
for the experiments in this study. The target infor-
mation depends on the questions we want to ask but
all of the input data for experiments are the number
of symptomatic cases recorded every day during the
simulation period, except for index date and next day.
For the prediction experiments, we split the entire
data into two groups, 80% for training and 20% for
testing. According to the target information we want
to predict, the answers are to be classified into differ-
ent numbers of levels. For example, in the experiment
of peak date prediction, there are roughly 360 levels
of target answers because there are 365 days in a year.
And to deal with the magnitude problem at the peak
value problem, we conduct a new method to catego-
rize the target information. The method is shown in
section 2.4, called variable length interval. The fur-
ther detail information about how we classified the
target answers is discussed in section 3. In order to
demonstrate the characteristics of our input data, we
take the level information of p
trans
to make compari-
son as it is the most important parameter among the
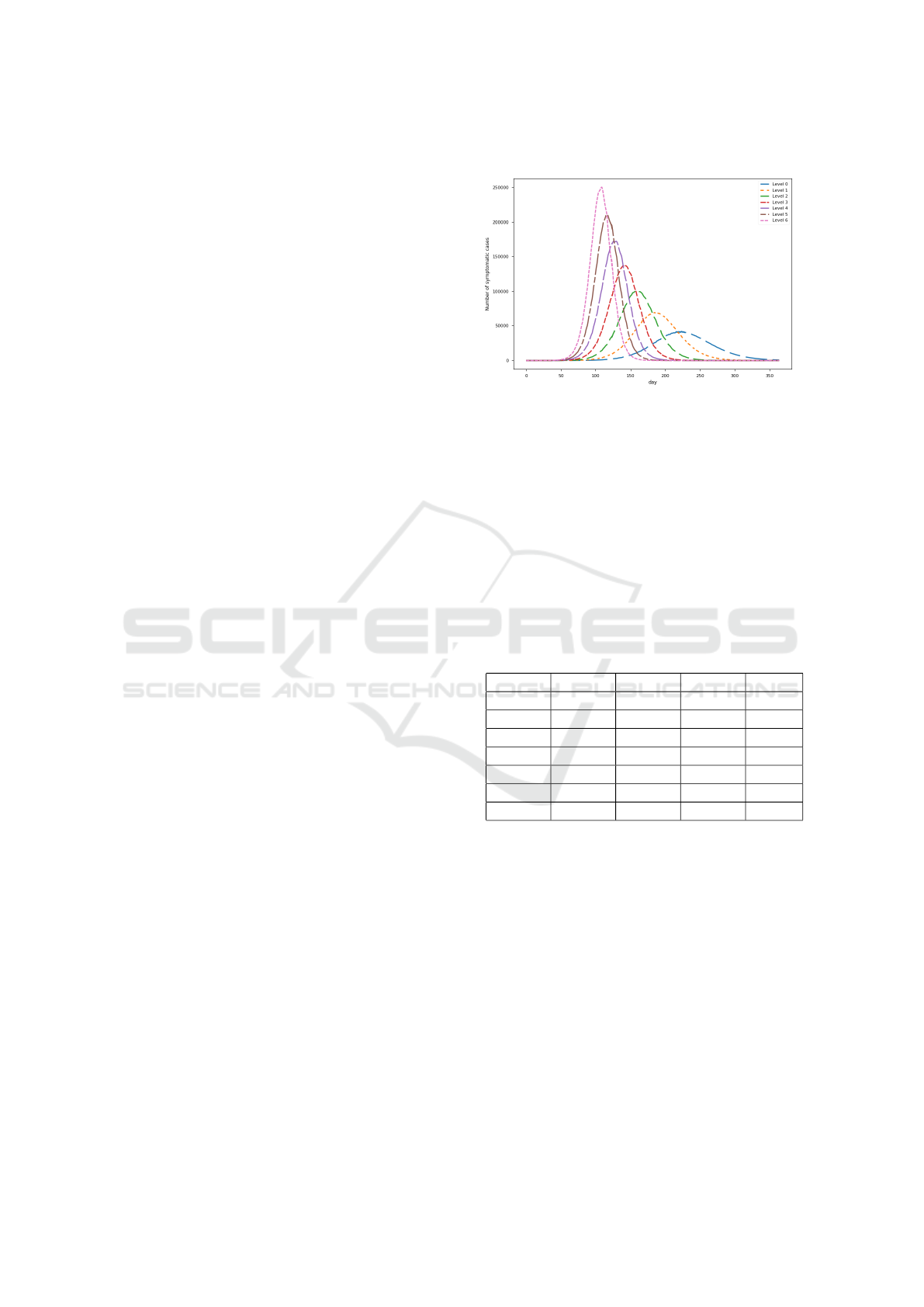
simulations as well as real world epidemics. In Figure
1, We show the average curve of the newly infected
symptomatic cases recorded every day for each level.
As the graph shown, the higher the level, the higher
as well as the earlier the peak is.
Figure 1: Epidemic curves.
In Table 1, we show the statistical characteristics
of the peak epidemics in each level. We can see that
the standard deviation of the peak value for each level
does not differ too much. However, the mean value
of the peak value varies significantly. In this table, Pr
99 and Pr 1 represent the value of the 99-percentile
and 1-percentile, respectively. The 1-percentile peak
value of level 0 and level 1 is 10 and 13 respectively.
We note that 10 index cases are created at the begin-
ning of the simulation and the peak value close to 10
actually means virus dose not spread far and the epi-
demic stops early.
Table 1: Level information.
Pr 99 Pr 1 Mean Std
Level 0 61239 10 50035 10921
Level 1 96981 13 81858 10968
Level 2 136102 98499 116361 10929
Level 3 177499 134424 158564 9795
Level 4 220326 169046 197707 11529
Level 5 263997 205040 239315 14079
Level 6 292110 237465 272069 11970
Below we provide a detailed account of data pre-
processing for each problem.
Data generation for predict p
trans
. The first ex-
periment, we feed the symptomatic cases of the whole
year to the neural network and let it predict the p
trans
level. In this task, the network got 98% accuracy on
the testing data set. Then we moved forward to see
if a good estimation can be achieved as early as pos-
sible. We, thus, only feed the training algorithm the
first few weeks, say from 2 weeks to 8 weeks, of sim-
ulation runs, and measure the performance.
Data generation for predict index case. We ran-
domly pick an interval of 49 days within the entire
epidemic and set the number of weeks when the index
case occurred before the interval of 49 days as the tar-
get answer. For example, if we choose the 49-day in-
Study Simulated Epidemics with Deep Learning
233
terval between the fourteenth day to the sixty-second
day of the entire epidemic as the input, the target an-
swer that we want to predict will be two, which means
the index case occurred two weeks before the 49-day
interval, that is, the first day of the entire epidemic.
The end of interval is restricted before the peak date
of each epidemic. If the peak date occurs at the first 49
days of the entire epidemic, we will simply pick the
first 49 days and the target answer will be zero. All of
the experiments of index date prediction use the fixed
data set in order to compare different results.
Data generation for the next day problem.
Instead of directly using the symptomatic cases
recorded every day, we use the difference between
two days as our input in this experiment, that is the
increased or decreased value of each day. If we con-
sider the unit of X-axis is day and Y-axis represents
the original value of symptomatic cases, it is just like
using the slope between two days as our input data to
feed into the network. The target information we want
to predict is also the increased or decreased value
compare to the final day of the known information.
In this task, we think the slope of two days may of-
fer more information than the raw value of each day
as the network can learn the increasing or decreasing
slope and get the correct direction of the next one. It
can also deal with the magnitude problem as the range
of the difference value is not that large compare to the
raw value of the symptomatic cases. So we can use
fixed interval here, which is set as 10 people for each
level. In the simulation world, we put the initial in-
fectious cases in the first day of the entire epidemic.
But in the real world, it is impossible to know the in-
dex date certainly. In this task, we first use the first
49-day interval as the input and predict the 50
th
day
of the epidemic. After getting an acceptable result,
we randomly move forward the starting point of the
49-day interval forward from 0 to 7 days, that is we
uniformly sample an integer from 0 to 7, and use the
sampled number as the starting point of our sequence.
The shifting of the starting point can be interpreted
as a test of the model’s tolerance level about the un-
certainty of the index date. We call such data shifted
data, and the outcome of learning with shifted data is
called the shifted case.
2.3 Deep Learning Architecture and
Experiment Setup
We use Stacked Long Short-Term Memory networks
launched by Keras(Keras, 2015) to set up our predic-
tion network, which is one of the most Recurrent Neu-
ral Networks. Unlike the traditional feed forward neu-
ral network, which can only deal with the current in-
put and output the corresponding results without con-
sidering past information, the design of Long Short-
Term Memory can help us record the previous infor-
mation and use it for the next round. The modules
of Long Short-Term Memory network have a loop to
reuse the previous information and a forget gate to de-
cide whether the information needs to be recorded or
not. It can record the information of last step as well
as the information far before it. Due to the ability of
combining previous information with the current in-
formation, the network is suitable for dealing with the
time series data, hence fits our purpose.
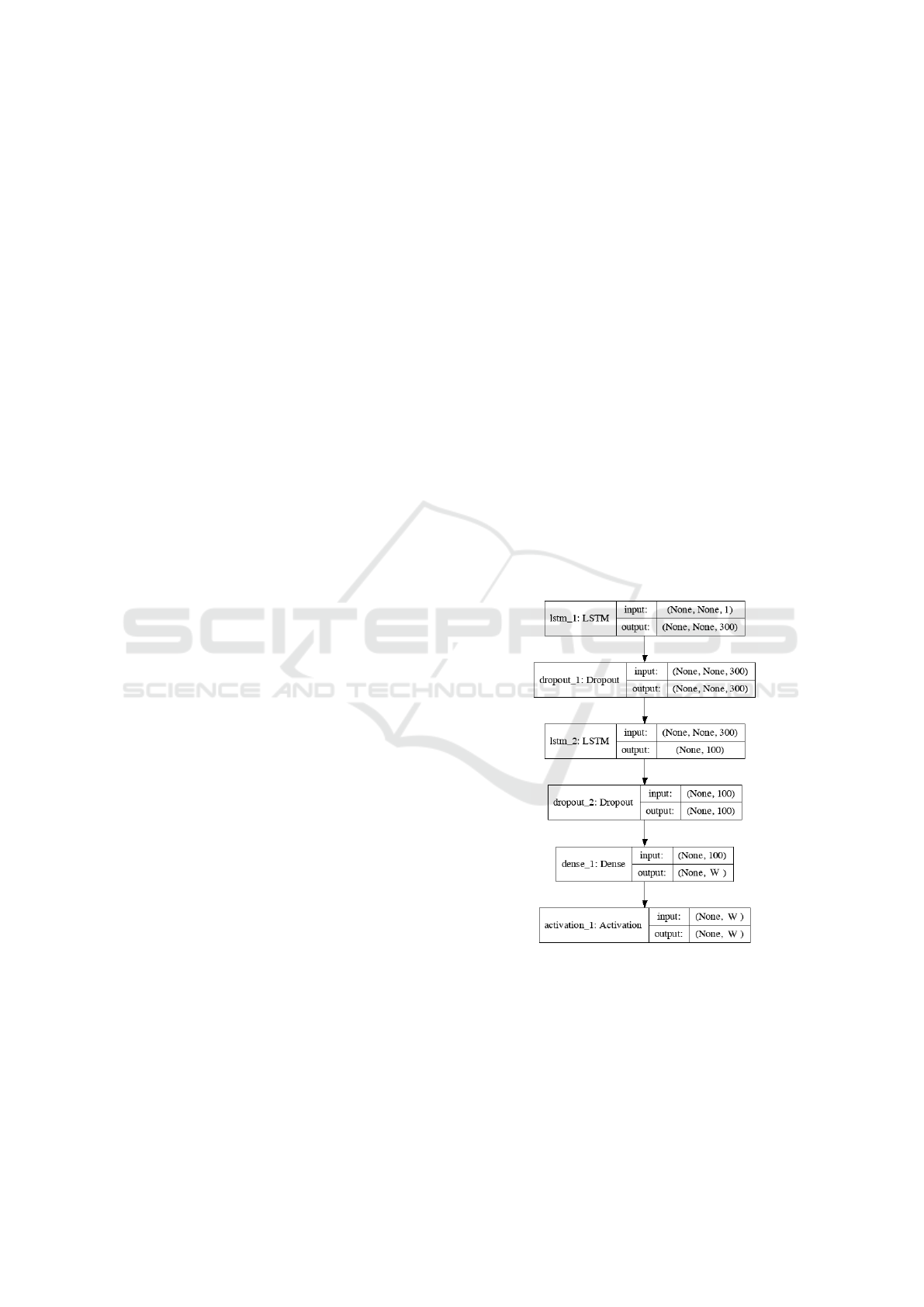
In Figure 2, we show the general network structure
of our experiments. The variable W depends on the
different number of target levels we want to predict
in each experiment. The dropout rate is set as 20%.
We use Adam as optimizer and categorical cross en-
tropy as loss function. The batch size is set as 32 and
the number of epochs is set around 15. The numbers
of parameters are about 362,000 in the first LSTM
layer, 160,000 in second LSTM layer, and 2,000 in the
dense layer respectively. The input and output in the
figure mean the dimension of input and output data of
each layer. All of the inputs fed into the model are the
infected cases.
Figure 2: Network structure.
In the study of epidemics, the severe and urgent
ones need most of the attention. Therefore, in our
study, we sometimes partition training data according
to its severity, that is p
trans
, to build models for each
severity level. We call this leveled training and de-
noted by Level x in the tables in section 3. On the
other hand, when we use training data of all levels to
build one model, the performance of this model facing
SIMULTECH 2019 - 9th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
234

different severity cases is also of interest. The testing
data is thus partitioned according to severity and we
get several testing results. We call this leveled testing
and use Lv x to denote a row is the result of leveled
testing with level x, this notation is only used at the
next day prediction problem.
2.4 Variable Length Interval
As the range of the peak value is too large, from ten
to about thirty thousand. If we directly transfer them
into categories and apply one hot encoding on them.
The target answer will be too sparse and the accuracy
will be really low. We also tried to predict the peak
value directly without any classification but it ended
up with the result of very large mean squared error
and the network learned nothing. For this task, if we
use fixed interval to convert the value, one hundred by
each level for example, it does not make sense as the
error of one hundred is different for the basic number
with two hundred and twenty thousand. So we con-
duct a method to convert the value of symptomatic
cases into several levels. We separate each level ac-
cording to the magnitude of the value. We want each
level to have a fixed percentile of the median value
in a level, not a fixed range of value for every levels.
The following is the formula of the conversion. The
range of the interval will become larger as the value
raises. And the median of each level is a geometric
series with the ratio of (1+α) / (1-α), the variable α is
the percentage of error we can accept. We set a basic
number β as the median of the first interval. And the
value below the basic number will be viewed as the
level 0. At this experiment, we set the basic number
β as one hundred and acceptable percentage α as 5%
and 10%.
Level = blog
(1+α)
(1−α)
PeakValue
β
+ 1.5c (1)
2.5 Measurements
To evaluate the utility of trained models, we have to
incorporate the requirements of real applications into
the measurements. For example, to predict the peak
date, it is acceptable to be off one or two days since
the peak date might be for the logistic planning or
speculating the potential impact on the healthcare de-
livery system. Therefore, a close enough prediction
is acceptable. Let L = {`
0
,...,`
k−1
} be the k set of
possible predictions. Let U = L × L → [0, 1] be the
utility function, and U(`
i
,`
j
) = 0.8 means that the
utility for the case that the model predicted `
i
while
the true case is `
j
is 0.8. Given a case with true label
`
i
and the prediction is (p
0
,..., p
k−1
), the utility is
∑
k−1
j=0
L( j,i)p( j). And the utility of a dataset is the av-
erage utility of each case in that dataset. The utility of
the model for a dataset is the expected utility, In this
paper, we use numerical values as symbols for `s, and
take the advantage that we can treat them as numer-
ical values so that arithmetic operations are possible.
Usually, the utility depends on the distance between
the predicted value and true value. For example, if the
predicted peak day is off by one day the utility is very
high or as good as the correct predictions. For those
situations, we can simplify the utility function to an
utility array U = (u
−k+1
,...,u
0
,...,u
k−1
) where the
subscript is the value of predicted index minus true
index. Therefore, u
0
is always 1. We use Acc ± 1
(Acc ±3) to denote the case that the immediate neigh-
bors(two neighbors) are considered correct prediction
respectively.
Given a value α, ∆
α
is the minimum d such that
the utility array with width 2d + 1 has utility no less
than α and use ∆
α
= ±d. In this paper, we use ∆0.9
and ∆0.95 to denote ∆
0.9
and ∆
0.95
when they appear
in the head row of a table.
When fixed length intervals are used for categoriz-
ing numerical data and when the range of the values
is large. The error in the absolute sense may not tell
the whole story. For each testing case, we define the
relative error as follows: re(`,tv) = |v(`) − tv|/v(`),
where ` is the predicted level, tv is the true value
and v() is a mapping from level to a numerical level,
here we choose the middle point of the interval. We
can then compute the mean relative error as the mean
value of the relative errors of all cases.
3 RESULTS
For each experiment, we first use the training data
of all levels to build the model for prediction. And
the result is identified as over all in the table below.
We then study the cases that given the fixed p
trans
level, how well the algorithms can learn. That is we
train seven different models individually, one for each
level. And we identify the results with the order of
level , that is the row level i in the table contains the
performance on each level. And all of the results re-
ported below comes from testing data, which has not
been seen by the model.
3.1 p
trans
In the experiments of p
trans
prediction, the dataset
contains fifty thousand records with p
trans
range from
0.075 to 0.105, divided into 7 levels.
Study Simulated Epidemics with Deep Learning
235
The retrospective estimation of p
trans
performs
well, the accuracy is higher than 98%, and the pre-
diction is off by at most one level. For the perspective
case, we carry out the experiment which uses first two
weeks data, that is to predict p
trans
after 2 weeks of
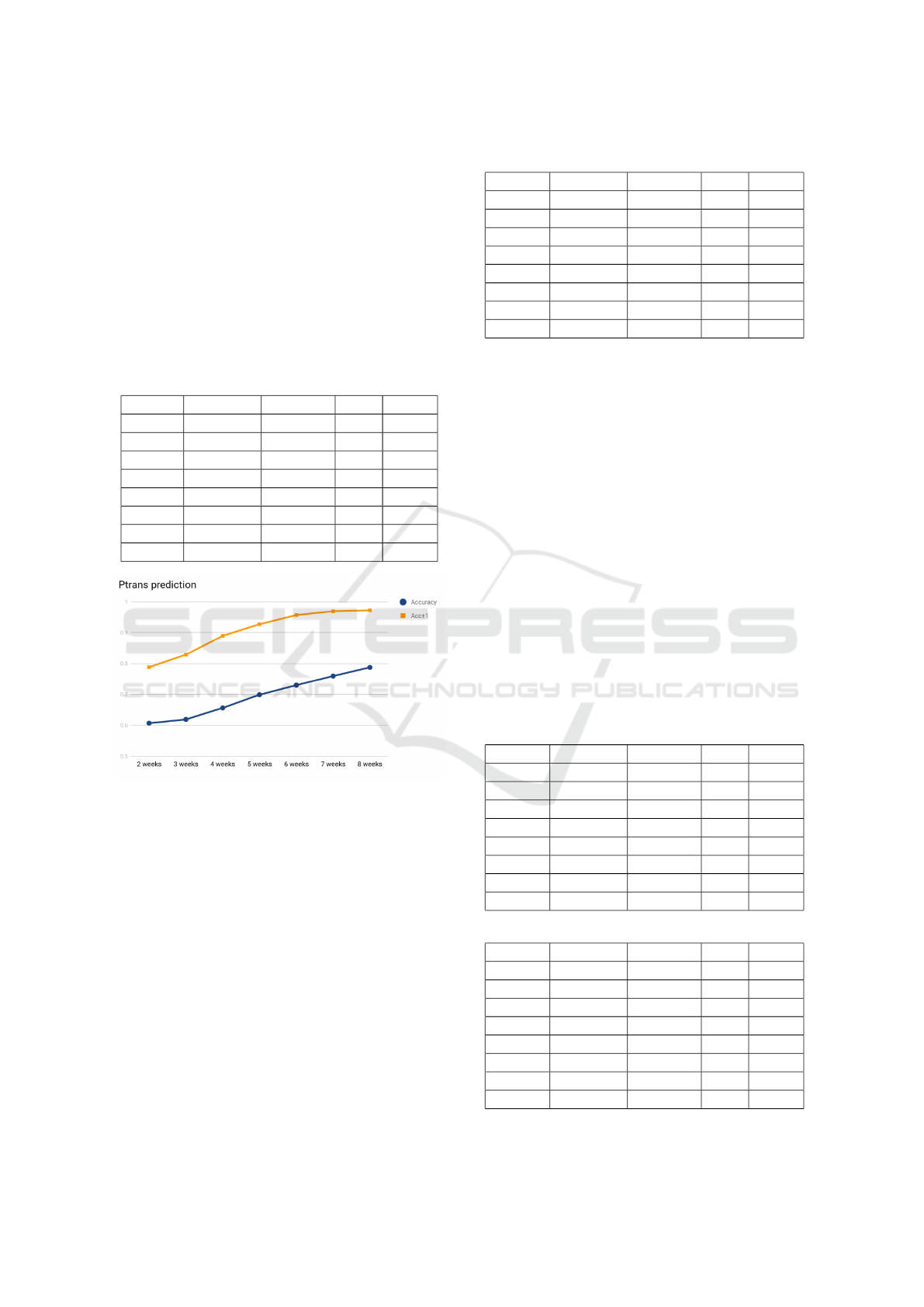
the epidemic, up to first eight weeks. As the results
shown in Table 2, two weeks data is enough to reach
over 60% accuracy and almost 80% of the cases is off
by one level and for the 8 week experiment, we have
over 97% accuracy within an error of one level. It is
worth noting that from 2 weeks to 8 weeks the growth
of the accuracy is almost a line as shown in Figure 3.
Table 2: Ptrans prediction.
Accuracy Acc±1 ∆0.9 ∆0.95
52 wks 98.471% 99.928% ±0 ±0
2 wks 60.685% 78.844% ±3 ±3
3 wks 61.908% 82.869% ±2 ±3
4 wks 65.627% 88.953% ±2 ±2
5 wks 69.907% 92.724% ±1 ±2
6 wks 72.995% 95.689% ±1 ±1
7 wks 75.940% 96.938% ±1 ±1
8 wks 78.742% 97.238% ±1 ±1
Figure 3: p
trans
prediction accuracy.
3.2 Peak Date
The unit for peak date prediction is day, that is each
level corresponds to one day. Although the overall
accuracy is only 50%, and ∆0.9 = ±17 as shown in
Table 3. However, we note that the prediction is done
at the end of the 7
th
week, which is quite early.
The results of level here is leveled learning as we
mentioned above. And the following level results are
the same as this experiment except for the next day
prediction experiment. We separate the data of each
level first then split the training and testing data to
train a model for each level. When we train different
models for different level of p
trans
, we note that in Ta-
ble 3 for higher level (level 3 to level 6) the prediction
is off by at most a week 95% of times.
Table 3: Peak date prediction.
Accuracy Acc±3 ∆0.9 ∆0.95
Level 0 57.919% 58.781% ±21 ±24
Level 1 60.636% 63.221% ±11 ±14
Level 2 63.949% 69.781% ±7 ±10
Level 3 63.287% 72.829% ±7 ±7
Level 4 63.660% 75.464% ±7 ±7
Level 5 68.522% 83.168% ±4 ±7
Level 6 62.698% 78.042% ±4 ±6
Overall 56.507% 63.232% ±17 ±28
3.3 Peak Value
The peak value is defined as the largest number of
people infected in one day for the entire epidemic pe-
riod. As mentioned above that we use variable length
interval to partition peak values. The benefit of it is
that this formulation corresponding to the relative er-
ror which suits our purpose better. However, one of
the drawback is the interpretation of the results need a
little more works. We carry out two experiments with
different variable length intervals, α = 0.1 (shown in
Table 4) and α = 0.05 (shown in Table 5) respectively.
It is expected that the accuracy for α = 0.05 case is
lower than α = 0.1 case, because it has smaller rela-
tive error. To be correct for the 0.05 case, the predic-
tion can not be off over 5%, while for 0.1 case it can
be off by as large as 10%. Roughly, for the 0.05 case,
Acc ± 1 = 0.96, means that 96% of predictions is off
by at most 15%. We can see that setting α to 0.05
produce better results at higher level.
Table 4: Peak value prediction α = 0.1.
Accuracy Acc±1 ∆0.9 ∆0.95
Level 0 70.377% 93.837% ±1 ±2
Level 1 76.011% 99.801% ±1 ±1
Level 2 79.722% 100% ±1 ±1
Level 3 76.739% 99.933% ±1 ±1
Level 4 84.350% 100% ±1 ±1
Level 5 84.625% 99.933% ±1 ±1
Level 6 81.084% 100% ±1 ±1
Overall 68.062% 88.994% ±2 ±2
Table 5: Peak value prediction α = 0.05.
Accuracy Acc±1 ∆0.9 ∆0.95
Level 0 61.962% 79.059% ±2 ±3
Level 1 63.029% 85.355% ±2 ±2
Level 2 62.624% 90.656% ±1 ±2
Level 3 66.402% 96.090% ±1 ±1
Level 4 70.225% 96.021% ±1 ±1
Level 5 71.239% 95.692% ±1 ±1
Level 6 75.661% 99.603% ±1 ±1
Overall 58.514% 75.461% ±3 ±5
SIMULTECH 2019 - 9th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
236
3.4 Infected Cases of Next Day
We try to study the possibility to build an online sys-
tem to predict the number of newly infected cases
based on the current situation. We first study the sim-
plest version of the problem, i.e., predict the outcome
of the 50
th
day given first 49 days. We call it fixed
case, since the first data item corresponding to a fixed
day, in this case first day, of the simulation. One way
to interpret the fixed case is that we know the index
date exactly. Next, we study the scenario that we are
not sure about the index date, but we know that index
date is in a time interval. In our experiment, for each
simulation run, we generate 7 sequences of length 49,
where the starting day ranged from the 1
st
day to the
7
th
day. This version is called shift window case.
Training starts with a preprocessing process, we
take the difference of the values of two consecutive
days to form a new sequence as the input to the learn-
ing algorithm, and the label is the difference between
next day and the last day in the input. The model
learns together with a postprocessing, that is adding
the value predicted with the value of last day in the
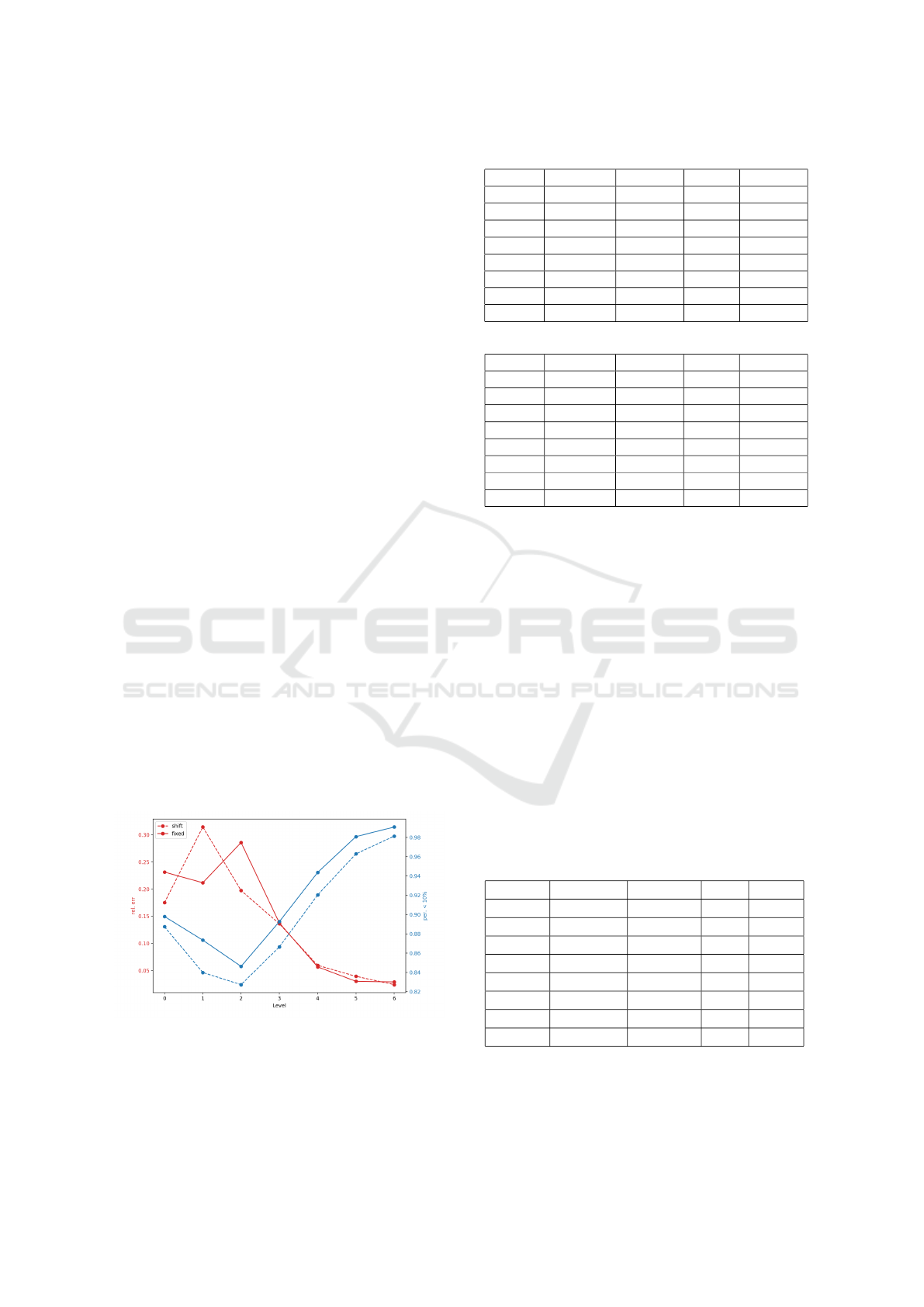
input. As shown in Figure 4, the accuracy and Acc±3
decrease as the testing level increase, the reason is that
we categorized the value with fixed length intervals.
When the level increases, a more infectious virus, the
newly infected cases also increase, thus more difficult
to predict the category correctly. Overall, the fixed
model performs better, except slightly higher mean
relative error as shown in Figure 4. This confirms our
intuition that the shift window version is harder than
the fixed one. Overall, the mean relative error is less
than 15% and almost 90% of the cases the prediction
is at most 10 % off the mark. We note that for more
severe epidemic, higher level ones, the mean relative
error is less than 6% as shown in Table 6 and 7.
Figure 4: Rel. err and per. below 10% of shift and fixed
data.
Table 6: Next day prediction (shift window).
Accuracy Acc±3 rel. err ≤10%
Lv 0 87.064% 96.054% 0.175 88.730%
Lv 1 76.521% 93.640% 0.314 83.987%
Lv 2 64.218% 89.874% 0.197 82.729%
Lv 3 56.685% 84.461% 0.136 86.647%
Lv 4 50.411% 75.920% 0.059 92.036%
Lv 5 48.045% 68.590% 0.039 96.290%
Lv 6 47.525% 61.999% 0.024 98.129%
Overall 62.570 % 83.005% 0.143 89.153%
Table 7: Next day prediction (fixed).
Accuracy Acc±3 rel. err ≤10%
Lv0 88.667% 96.288% 0.231 89.794%
Lv1 81.908% 94.102% 0.211 87.342%
Lv2 68.986% 92.710% 0.285 84.625%
Lv3 60.569% 89.993% 0.137 89.264%
Lv4 55.238% 83.289% 0.056 94.363%
Lv5 52.087% 76.076% 0.030 98.078%
Lv6 51.455% 70.105% 0.029 99.074%
Overall 66.642% 87.307% 0.148 91.232%
3.5 Index Date
Last, we study the problem that given a segment of
an epidemic, predicting how far is the first day in the
sequence from the first date of the epidemic. And we
set the unit of prediction to be week. As show in Ta-
ble 8, the over all accuracy is not impressive although
we already set the predicting unit to week, instead
of day in the case for predicting peak date. We do
note a recurrent scenario also show in Table 8, that
the model trained with higher level of p
trans
performs
better than lower level cases. Although, it is too early
to claim that this is a difficult task to build a prediction
model. We do want to mention that a negative result
with simulated data, can be seen as a indication to the
difficulty of the problem in real world. Because the
real world data is much more chaotic with infinitely
many sources of noises.
Table 8: Index date prediction.
Accuracy Acc±1 ∆0.9 ∆0.95
Level 0 24.254% 49.901% ±5 ±7
Level 1 26.508% 64.016% ±3 ±4
Level 2 38.436% 78.794% ±2 ±3
Level 3 52.154% 88.867% ±2 ±2
Level 4 60.146% 90.981% ±1 ±2
Level 5 66.534% 96.156% ±1 ±1
Level 6 70.504% 96.825% ±1 ±1
Overall 39.397% 69.509% ±4 ±7
Study Simulated Epidemics with Deep Learning
237

4 CONCLUSION AND FUTURE
WORKS
The preliminary results are promising. It is worth
pointing out that when the infectiousness is low, i.e.,
the p
trans
at lower level, the disease control agency
only has to monitor its progress, when the infectious
is very high, there is not much the agency can do.
Therefore, the interesting cases are those in the mid-
dle. We note that the prediction is more accurate with
higher level infectiousness. Also our choice of the
utility functions more or less reflect the real applica-
tions. For the prediction of peak value, we do not
use fix length partition, which corresponding to the
idea of absolute error. Instead, we use variable length
interval, which is corresponding to relative error. We
believe this trick can be applied elsewhere. When pre-
dicting next day, we feed the model with the sequence
of the differences between two consecutive days, this
corresponding to take the derivative of the epidemic
curve at given day. This is also an interesting trick
which might be useful in other situations. We note
that the deep learning performed not so well for pre-
dicting the index date. One possible interpretation is
that this is really a difficult problem even in the sim-
plified simulated world. The hope to get a good esti-
mation in the real world might even be more difficult.
Therefore, a not so positive result can shed light on the
limitation of what can be learnt in real world and we
might want to frame the problem differently to hope
for better results.
We plan to try other machine learning approaches,
especially regression based ones like SVR so that one
might get better understanding about the capacity and
limitation of deep learning methods on simulated epi-
demiology data. From disease control perspective,
one obvious future direction is to include mitigation
strategies, such as vaccination, and social distancing
so that the outcome of various combination of miti-
gation can be learned. The parameter spaces will be
much larger when mitigation strategies included, and
the power of deep learning can be further explored.
To use more detailed information of a simulated epi-
demic, such as geographic location is also an inter-
esting and important next step. Furthermore, how to
apply the trained model in the real situation, is also a
very important yet challenging problem. We can start
by introducing additive random noise to the output of
simulation system to mimic the background or base-
line disease states in real world and feed the perturbed
data into the learning system. One exciting idea is
to use the generative adversarial model (Goodfellow
et al., ) to train a generative model to generate simu-
lation results without running the simulation.
ACKNOWLEDGEMENTS
This study is supported in part by MOST, Tai-
wan, Grant No. MOST107-2221-E-001-017-MY2
and MOST107-2221-E-001-005 and by Multidisci-
plinary Health Cloud Research Program: Technology
Development and Application of Big Health Data,
Academia Sinica, Taipei, Taiwan.
REFERENCES
Anderson, R. and May, R. (1992). Infectious Diseases of
Humans: Dynamics and Control. Dynamics and Con-
trol. OUP Oxford.
Chang, H.-J., Chuang, J.-H., Fu, Y.-C., Hsu, T.-S., Hsueh,
C.-W., Tsai, S.-C., and Wang, D.-W. (2015). The im-
pact of household structures on pandemic influenza
vaccination priority. In SIMULTECH2015, pages
482–487.
Fu, Y.-c., Wang, D.-W., and Chuang, J.-H. (2012). Rep-
resentative contact diaries for modeling the spread of
infectious diseases in taiwan. PLoS ONE, 7(10):1–7.
Germann, T. C., Kadau, K., Longini, I. M., and Macken,
C. A. (2006). Mitigation strategies for pandemic in-
fluenza in the United States. Proceedings of the Na-
tional Academy of Sciences, 103(15):5935–5940.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B.,
Warde-Farley, D., Ozair, S., Courville, A., and Ben-
gio, Y. Generative adversarial nets. In Advances in
Neural Information Processing Systems 27.
Keras (2015). Keras documentation. https://keras.io/.
Li, M. and Vitnyi, P. M. (2008). An Introduction to Kol-
mogorov Complexity and Its Applications. Springer.
Mnih, V., Kavukcuoglu, K., Silver, D., Graves, A.,
Antonoglou, I., Wierstra, D., and Riedmiller, M. A.
(2013). Playing atari with deep reinforcement learn-
ing. CoRR, abs/1312.5602.
Tsai, M.-T., Chern, T.-C., Chuang, J.-H., Hsueh, C.-W.,
Kuo, H.-S., Liau, C.-J., Riley, S., Shen, B.-J., Shen,
C.-H., Wang, D.-W., and Hsu, T.-S. (2010). Efficient
simulation of the spatial transmission dynamics of in-
fluenza. PLOS ONE, 5(11):1–8.
SIMULTECH 2019 - 9th International Conference on Simulation and Modeling Methodologies, Technologies and Applications
238
