
Calibration Techniques for Binary Classification Problems: A
Comparative Analysis
Alessio Martino
a
, Enrico De Santis
b
, Luca Baldini
c
and Antonello Rizzi
d
Department of Information Engineering, Electronics and Telecommunications, University of Rome ”La Sapienza”,
Via Eudossiana 18, 00184 Rome, Italy
Keywords:
Calibration, Classification, Supervised Learning, Support Vector Machine, Probability Estimates.
Abstract:
Calibrating a classification system consists in transforming the output scores, which somehow state the con-
fidence of the classifier regarding the predicted output, into proper probability estimates. Having a well-
calibrated classifier has a non-negligible impact on many real-world applications, for example decision mak-
ing systems synthesis for anomaly detection/fault prediction. In such industrial scenarios, risk assessment is
certainly related to costs which must be covered. In this paper we review three state-of-the-art calibration
techniques (Platt’s Scaling, Isotonic Regression and SplineCalib) and we propose three lightweight proce-
dures based on a plain fitting of the reliability diagram. Computational results show that the three proposed
techniques have comparable performances with respect to the three state-of-the-art approaches.
1 INTRODUCTION
Classification is one of the most important problems
falling under the machine learning and, specifically,
under the supervised learning umbrella. Generally
speaking, it is possible to sketch three main families:
clustering, regression/function approximation, classi-
fication. These problems mainly differ on the nature
of the process to be modelled by the learning system
(Martino et al., 2018a).
More into details, let P : X → Y be an orientated
process from the input space X (domain) towards the
output space Y (codomain) and let hx,yi be a generic
input-output pair drawn from P, that is y = P(x).
In supervised learning a finite set S = hX,Y i of
input-output pairs is supposed to be known and com-
mon supervised learning tasks can be divided in clas-
sification and function approximation. In the former
case, the output space Y is a non-normed space and
output values usually belong to a finite categorical set
of possible values. Conversely, in the latter case, the
output space is a normed space (usually R). In unsu-
pervised learning there are no output values and reg-
ularities have to be discovered using only informa-
a
https://orcid.org/0000-0003-1730-5436
b
https://orcid.org/0000-0003-4915-0723
c
https://orcid.org/0000-0003-4391-2598
d
https://orcid.org/0000-0001-8244-0015
tion from X . The seminal example is data clustering,
where aim of the learning system is to return groups
(clusters) of data in such a way that patterns belong-
ing to the same cluster are more similar with respect to
patterns belonging to other clusters (Jain et al., 1999;
Martino et al., 2017b; Martino et al., 2018b; Martino
et al., 2019; Di Noia et al., 2019).
Synthesizing a classifier (predictive model) con-
sists in feeding some hx,yi pairs to a training algo-
rithm in such a way to automatically learn the under-
lying model structure. In other words, the classifier
learns a decision function f that, given an input x, re-
turns a predicted class label ˆy, i.e. a prediction regard-
ing the class that pattern may belong to:
ˆy = f (x) (1)
Eq. (1) is usually referred to as hard classifica-
tion. Probabilistic classifiers can also return a pos-
terior probability P(output|input) which can be use-
ful for many real-world applications, for example
condition-based maintenance, decision support sys-
tems or anomaly/fault detection as operators usually
want to know the probability of a specific equipment
to fail given some input (known) state/conditions
(De Santis et al., 2018b). Trivially, probabilistic clas-
sifiers can be ’forced’ to return hard predictions by
letting
ˆy = argmax
y
P(Y = y|X) (2)
Martino, A., De Santis, E., Baldini, L. and Rizzi, A.
Calibration Techniques for Binary Classification Problems: A Comparative Analysis.
DOI: 10.5220/0008165504870495
In Proceedings of the 11th International Joint Conference on Computational Intelligence (IJCCI 2019), pages 487-495
ISBN: 978-989-758-384-1
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
487

that is, for a given input pattern x ∈ X, the classifier
assigns the output label y ∈ Y which corresponds to
the maximum posterior probability.
Albeit not all classifiers are probabilistic classi-
fiers, some classifiers such as Support Vector Machine
(SVM) (Boser et al., 1992; Cortes and Vapnik, 1995;
Sch
¨
olkopf and Smola, 2002; Cristianini and Shawe-
Taylor, 2000) or Na
¨
ıve Bayes may return a score s(x)
which somewhat states the ’confidence’ in the predic-
tion of a given pattern x. As regards Na
¨
ıve Bayes, this
score can be seen as the probability estimate for class
membership. However, this score is not calibrated
(Domingos and Pazzani, 1996). For SVMs, the score
is basically the distance with respect to the separat-
ing hyperplane: the sign of s(x) determines whether
x has been classified as positive or negative, whereas
the magnitude of s(x) determines the distance with re-
spect to the hyperplane. Conversely to Na
¨
ıve Bayes,
SVMs’ scores not only are not calibrated, but also are
not bounded in [0, 1], albeit some re-scaling can be
performed (Zadrozny and Elkan, 2002).
Formally speaking, a classifier is said to be well-
calibrated if P(y|s(x) = s), that is, the probability for
a pattern x to belong to a label y converges to the
score s(x) = s as the number of samples tends to infin-
ity (Murphy and Winkler, 1977; Zadrozny and Elkan,
2002). In plain terms, the calibration of a classifica-
tion system consists in mapping the scores (or not-
calibrated probability estimates) into proper probabil-
ity estimates bounded in range [0,1] by definition.
The aim of this paper is to investigate amongst
several calibration techniques by considering binary
classification problems using SVM as classification
system. The remainder of this paper is structured as
follows: in Section 2 we give an overview of exist-
ing calibration techniques and figures of merit for ad-
dressing the goodness of the calibration along with
three new lightweight procedures to be compared
with state-of-the-art approaches; in Section 3 we de-
scribe the datasets used for experiments, along with
comparative results amongst the considered methods;
Section 4 concludes the paper, suggesting future re-
search and applications.
2 AN OVERVIEW OF
CALIBRATION TECHNIQUES
2.1 Current Approaches
In order to quantify the calibration of a given classifier
the reliability diagram is usually employed (Murphy
and Winkler, 1977). The reliability diagram is built as
follows:
• scores/probabilities go on the x-axis
• empirical probabilities P(y|s(x) = s), namely the
ratio between the number of patterns in class y
with score s and the total number of patterns with
score s, go on the y-axis
and if the classifier is well-calibrated, then all points
lie on the y = x line (i.e., the scores are equal to the
empirical probabilities). In case of binary classifi-
cation, the empirical probabilities regard the positive
instances only (i.e., the ratio between the number of
positive instances having score s and the total number
of instances with score s).
Since scores are normally real-valued scalars, it is
quite impossible to quantify the number of data points
sharing the same score
1
. In this case, a binning pro-
cedure is needed:
• on the x-axis, the average score value within the
bin is considered
• on the y-axis, we get the ratio between the number
of patterns in class y lying in a given bin and the
total number of patterns lying in the same bin.
In works such as (Zadrozny and Elkan, 2002) and
(Niculescu-Mizil and Caruana, 2005) the authors pro-
posed to consider 10 equally-spaced bins in range
[0,1], regardless of the distribution of the scores
within that range. For some datasets, however, this
might not be a good choice and suitable alternatives
which somewhat consider the available samples are:
• The Scott’s rule (Scott, 1979) evaluates the bin
width according to the number of samples (scores)
n and their standard deviation σ as
bin witdh =
3.5 ·σ
n
1/3
• The Freedman–Diaconis rule (Freedman and Dia-
conis, 1981) evaluates the bin width as follows
bin witdh =
2 ·IQR
n
1/3
where IQR is the inter-quantile range
• The Sturges’ formula (Sturges, 1926) evaluates
the number of bins as follows
number of bins = 1 + dlog
2
ne
where d·e denotes the ceiling function
• The square root choice, where the number of bins
is given by
number of bins =
√
n
1
This counting procedure will return the (trivial) value
of 1 for any s(x).
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
488

However, using a single binning, even if evaluated ac-
cording to one of the four alternatives above, might
not be a good choice, especially if data do not fol-
low a specific underlying distribution (e.g., uniform
distribution in case of uniform binning or normal dis-
tribution in case of the Sturges’s formula). To this
end, in (Naeini et al., 2015), the Authors proposed
the Bayesian Binning into Quantiles technique, which
considers different binning (and their combination) in
order to make the calibration procedure more robust.
Let c denote the positive class and let us assume
P(c|x) = 1 for positive patterns and P(c|x) = 0 other-
wise. After training a classifier such as SVM, aim of
the calibration procedure is to find a function f for-
mally defined as
f : s(x) →
ˆ
P(c|x) (3)
hence, in other words, a function (model) in charge of
transforming score values into probability estimates.
One of the most famous techniques is the Platt’s
scaling (Platt, 2000; Niculescu-Mizil and Caruana,
2005), a parametric approach in order to estimate
P(y = 1|s(x)), namely the probability that a given pat-
tern x belongs to the positive class. Platt’s discussion
starts by using the Bayes’ formula
P(y = 1|s(x)) =
p(s(x)|y = 1)P(y = 1)
∑
i={±1}
p(s(x)|y = i)P(y = i)
(4)
where P(y = i) are prior probabilities and p(s(x)|y =
i) are the class conditional densities (i.e., the prob-
ability density function for belonging to class i). In
order to use Eq. (4), one can estimate the class con-
ditional densities by considering the normalized his-
tograms of the scores as returned by the SVM. Platt
showed that if the margin between the histograms of
the two classes have an exponential trend, then Bayes’
rule leads to
P(y = 1|s(x)) =
1
1 + exp{As(x) + B}
(5)
which is a plain parametric sigmoid function and tun-
ing the calibration model basically consists in find-
ing the two parameters A and B. Platt suggests to
minimize the negative log-likelihood on some train-
ing data by means of a model-trust optimization pro-
cedure based on the Levenberg-Marquardt algorithm.
In (Lin et al., 2007) an improved optimization proce-
dure based on Newton’s method is proposed. Platt’s
scaling has been proved to be successful if the relia-
bility diagram of the dataset shows a sigmoidal trend.
An alternative technique relies on isotonic regres-
sion (Zadrozny and Elkan, 2002; Zadrozny and Elkan,
2001). Pair-Adjacent Violators (Ayer et al., 1955) is
one of the main algorithms in order to compute an iso-
tonic regression. Given a real-valued vector x ∈ R
n
and a weights vector
2
w ∈ R
n
such that x
i
≥ x
i−1
and
w
i
> 0 for all i = 1,.. .,n, then the isotonic regression
of a function f (x) consists in finding a function g(x)
according to a mean squared error criterion
n
∑
i=1
w
i
(g(x
i
) − f (x
i
))
2
(6)
where g(x) must be a piecewise non-decreasing (iso-
tonic) function. By letting y and s be the vectors
containing the output class labels and their respective
scores, Pair-Adjacent Violators works as follows:
1. sort y according to s: if y is already isotonic
3
, then
return the estimate
ˆ
y ≡ y, otherwise initialize the
estimate values as
ˆ
y = y
2. if
ˆ
y is not isotonic, there must exist an index i such
that
ˆ
y
i
≤
ˆ
y
i−1
: for these values
4
we estimate
ˆ
y
i
=
ˆ
y
i−1
=
ˆ
y
i
+
ˆ
y
i−1
2
3. repeat step 2 until
ˆ
y is isotonic.
At the end of this procedure,
ˆ
y contains ordered values
(probability estimates) for scores in s. Further, due
to the piecewise nature of isotonic regression,
ˆ
y will
contain few different values, each of which is repeated
several times. Generally, Pair-Adjacent Violators re-
turns more samples in the score space where patterns
have been misclassified and less samples where pat-
terns have been correctly classified.
A recently proposed method is called SplineCalib
(Lucena, 2018) which aims at overcoming the major
drawbacks of Platt’s scaling and isotonic regression:
• the Platt’s scaling is based on the empirical ob-
servation that the relationship between scores and
probabilities are often well-fitted by a sigmoid
function: obviously, this works well only when
the data fit the model, but performs poorly when
the calibration function is not well-approximated
by a sigmoid function
• the Platt’s scaling works well for few calibration
data (less than 1000 instances), but the isotonic
regression overcomes this limitation
• the nature of piecewise constant approximation
given by isotonic regression opens to a wider fam-
ily of calibration function; however, its coarseness
can be a drawback.
2
This most general form is usually referred to as
weighted isotonic regression. However, in this work, the
weights vector is omitted.
3
All 0’s followed by all 1’s since we are considering bi-
nary classification.
4
The properly-said ”pair-adjacent violators” since they
violate the isotonic trend.
Calibration Techniques for Binary Classification Problems: A Comparative Analysis
489

As its name suggests, SplineCalib is based on (cu-
bic) smoothing splines. Like isotonic regression,
SplineCalib is a non-parametric approach and, at the
same time, unlike isotonic regression, SplineCalib fits
a cubic spline instead of a piecewise constant approx-
imation. In standard spline interpolation, one chooses
a set of knots and fits a polynomial (usually with de-
gree 3 or 4) within each interval: the more knots, the
better the fitting of the data but also high risk of over-
fitting. Smoothing splines (Wahba, 1990) may also
use all of the available points as knots and perform
a regularized penalty on the second derivative of the
function. Given a relationship between predictors x
and output y of the form y = f (x), the smoothing
spline estimate
ˆ
f of f is the function, amongst the
twice-differentiable ones, that minimize
n
∑
i=1
y
i
−
ˆ
f (x
i
)
2
+ λ
Z
ˆ
f
00
(t)dt (7)
In (Lucena, 2018), instead of minimizing the sum
of squares, the Author proposes to employ a log-
likelihood criterion instead
−
n
∑
i=1
(y
i
·log
ˆ
f (x
i
)) + (1 −y
i
) ·log(1 −
ˆ
f (x
i
))
+
+
1
2
λ
Z
ˆ
f
00
(t)dt (8)
which resembles logistic regression (Hastie et al.,
2001). Both Eqs. (7) and (8) see the regularization
term λ ≥ 0 which weights the contribution between
goodness of fit (leftmost term) and roughness (right-
most term). Specifically, if λ →0 no smoothing is tol-
erated, with risk of overfitting; conversely, if λ → ∞
no curvature is tolerated, with risk of going towards
an ordinary least squares interpolation.
The probability estimates via SplineCalib can be eval-
uated by the following steps:
1. sample K knots
5
from the unique items in the
score vector s
2. build the natural basis expansion matrix X ∈R
n×K
between values in s and the K knots. Given a set
of ordered knots {φ
1
,.. .,φ
K
}, the natural cubic
spline basis is defined as
N
1
(x) = 1
N
2
(x) = x (9)
N
k+2
(x) = d
k
(x) −d
K−1
(x) ∀k = 1,... ,K −2
where d
k
=
(x−φ
k
)
3
+
−(x−φ
K
)
3
+
φ
K
−φ
k
3. perform an `
2
-regularized logistic regression over
the pair hX,yi by considering a candidate value
5
One can also use all the available points, yet the Author
states that 200 points suffice.
set for λ and choose the best value, say λ
?
, as the
one that returns the best cross-validation log-loss
4. re-fit hX,yi using λ
?
5. return the calibration function f (s) by composing
the basis expansion of s and the fitted model from
the previous step in order to return the probability
estimate.
2.2 Proposed Techniques
All of the three methods explained so far share the
common goal to properly fit the reliability diagram:
the better the fit, the more reliable the resulting prob-
ability estimates. Whilst the three methods use the
’score–label’ pairs in order to accomplish this task, we
investigate an alternative exercise by fitting the points
lying on the reliability diagram. Hence, instead of
working with ’score–label’ pairs, we work with ’aver-
age bin value–fraction of positive patterns in that bin’
pairs. As will be clear in Section 3, a reliability dia-
gram almost never shows a linear trend, hence this fit-
ting shall rely on more sophisticated functions
6
. For
these exercises we use:
1. polynomial fitting: the points lying on the relia-
bility diagram are fitted by means of 3-degree and
4-degree polynomials
2. spline fitting: after choosing a suitable number of
knots and considering the corresponding intervals,
within each interval a natural cubic spline interpo-
lation is performed.
2.3 Figures of Merit
It is important to quantify the goodness of the calibra-
tion, hence how the probability estimates are far from
the empirical probabilities. To this end, two meth-
ods have been proposed in literature: the Brier score
(Brier, 1950; DeGroot and Fienberg, 1983) and the
Log-Loss score.
Given a series of N known events and the respec-
tive probability estimates, the Brier score is the mean
squared error between the outcome o (1 if the event
has been verified and 0 otherwise) and the probability
p ∈ [0,1] assigned to such event. Hence, in its most
general form, the Brier score has the form:
BS =
1
N
N
∑
i=1
(o
i
−p
i
)
2
(10)
6
Indeed, none of the methods introduced so far (Platt’s
scaling, isotonic regression, SplineCalib) use a linear fitting.
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
490

In the context of binary classification, Eq. (10) can be
specifically written as:
BS =
1
N
N
∑
i=1
(T (y
i
= 1|x
i
) −P(y
i
= 1|x
i
))
2
(11)
where T (y
i
= 1|x
i
) = 1 if y
i
= 1 and T (y
i
= 1|x
i
) = 0
otherwise and P(y
i
= 1|x
i
) is the estimated probability
for pattern x
i
to belong to class 1. As the Brier score
resembles the mean squared error, a lower value is
preferred.
The Log-Loss for binary classification is defined
as follows:
LL = −
1
N
N
∑
i=1
[y
i
logp
i
+ (1 −y
i
)log(1 − p
i
)] (12)
and, as per the Brier score, the lower, the better.
The Log-Loss index matches the estimated probabil-
ity with the class label with logarithmic penalty: for
small deviations between y
i
and p
i
the penalty is low,
whereas for large deviations the penalty is high.
3 TEST AND RESULTS
3.1 Datasets Description
For addressing the calibration performances of the
three state-of-the-art methods, namely Platt’s scaling
(PS), isotonic regression (IR), SplineCalib (SC) and
the three fitting methods from Section 2.2, namely
3-degree polynomial (Poly3), 4-degree polynomial
(Poly4) and natural cubic spline (NCS), two bench-
mark datasets from the UCI Machine Learning repos-
itory (Dua and Graff, 2019) have been considered:
Adult: the ADULT dataset is composed by 48842 in-
stances and 14 attributes and the goal is to predict
whether a person earns more than 50000$ per year
based on census data
Abalone: the ABALONE dataset is composed by
4177 instances and 8 attributes and the goal is
to predict the age of abalone from physics mea-
surements. Since the Abalone dataset is natively
multiclass (or for regression problems), we con-
sidered the median age and all output values be-
low the median have been marked as 1 and the
remaining values have been marked as 0.
These two datasets are freely available and have been
extensively used as benchmarks for a plethora of
learning techniques. Further, ADULT has been used
in all major works on calibration techniques, see
(Zadrozny and Elkan, 2002) for IR, (Platt, 2000) for
PS and (Lucena, 2018) for SC. Alongside these two
benchmark datasets, an additional dataset (hereinafter
PCN) has been considered as well, where aim of the
classification system is to predict whether a protein is
an enzyme or not. This is a striking example of real-
world problem in which a good probability estima-
tion plays a huge role (Minneci et al., 2013; Li et al.,
2016). The 3-dimensional folded structure of a pro-
tein can be described by its Protein Contact Network
(Di Paola et al., 2012), an undirected and unweighted
graph where nodes correspond to residues’ α-carbon
atoms and edges are scored if the Euclidean distance
between nodes’ spatial arrangements is within [4,8]
˚
A.
However, proteins notably have different sizes and
some pre-processing stages need to be performed in
order to map graphs into real-valued vectors of the
same length. Following (Maiorino et al., 2017) and
(Martino et al., 2017a), let A and D be the adjacency
and degree matrices for a given graph G. The Lapla-
cian matrix L is defined as
L = D −A (13)
and the normalized Laplacian matrix L reads as
L = D
−1/2
LD
−1/2
(14)
If the graph G has m nodes, then A,D, L,L ∈ R
m×m
and none of these matrices can directly be used in
order to properly match two graphs having different
sizes. In order to overcome this problem, we consider
the following property (Butler, 2016): the eigenval-
ues of L lie in range [0,2] regardless of the underlying
graph. However, the number of eigenvalues equals the
number of nodes m, hence neither the spectrum of L
can be used in order to compare two graphs. The fi-
nal step is to consider the spectral density of the graph
G by using a kernel density estimator (Parzen, 1962)
with Gaussian kernel. Given Λ = {Λ
1
,.. .,Λ
m
} as the
spectrum of L, the corresponding graph spectral den-
sity can be evaluated as
p(x) =
1
m
m
∑
i=1
1
√
2πσ
2
exp
−(x −Λ
i
)
2
2σ
2
(15)
where σ determines the kernel bandwidth and in order
to consider a suitable value that scales in a graph-wise
fashion, we used the Scott’s rule (cf. Section 2.1).
The distance between two graphs, say G
1
and G
2
, can
be evaluated as the `
2
norm between their respective
spectral densities, say p
1
(x) and p
2
(x):
d(G
1
,G
2
) =
Z
2
0
(p
1
(x) − p
2
(x))
2
dx (16)
Finally, 100 samples linearly spaced in [0,2] have
been extracted from the density function evaluated
with Eq. (15). Such final 100 samples unambiguously
identify each graph which, to this stage, is a vector
Calibration Techniques for Binary Classification Problems: A Comparative Analysis
491

in R
100
and in turn the dissimilarity measure between
patterns, formerly Eq. (16), collapses into the plain
Euclidean distance. This preprocessing stage has
been performed on a subset of the Escherichia coli
str. K12 proteome. Initially, the entire proteome gath-
ered from UniProt (The UniProt Consortium, 2017)
has been considered. After cross-checking with the
Protein Data Bank database (Berman et al., 2000),
all unresolved proteins have been removed. Fur-
ther, in order to consider only good quality and reli-
able atomic coordinates, proteins with no information
about the measurement resolution and proteins whose
measurement resolution is greater than 3
˚
A have been
removed. Networks with at least one isolated node
have not been considered either since it is impossible
to evaluate Eq. (14). Finally, very few large protein
complexes with over 2000 nodes have been removed
as well. These filtering procedures returned a total
number of 6061 proteins which have been labelled 1
if they have been assigned to an Enzyme Commis-
sion number (Webb, 1992), so they show enzymatic
properties, and 0 otherwise. Subsets of this dataset
have already been analyzed in works such as (Mar-
tino et al., 2017a; De Santis et al., 2018a; Martino
et al., 2018c).
3.2 Comparative Results
The three datasets (ADULT, ABALONE and PCN) have
been split into training (70% of the available patterns)
and test set (the remaining 30%). For all datasets a
3-fold cross-validation has been performed for hyper-
parameters tuning and model calibration tuning. For
all experiments, we considered a SVM classifier be-
cause it is a well-known uncalibrated binary classifier.
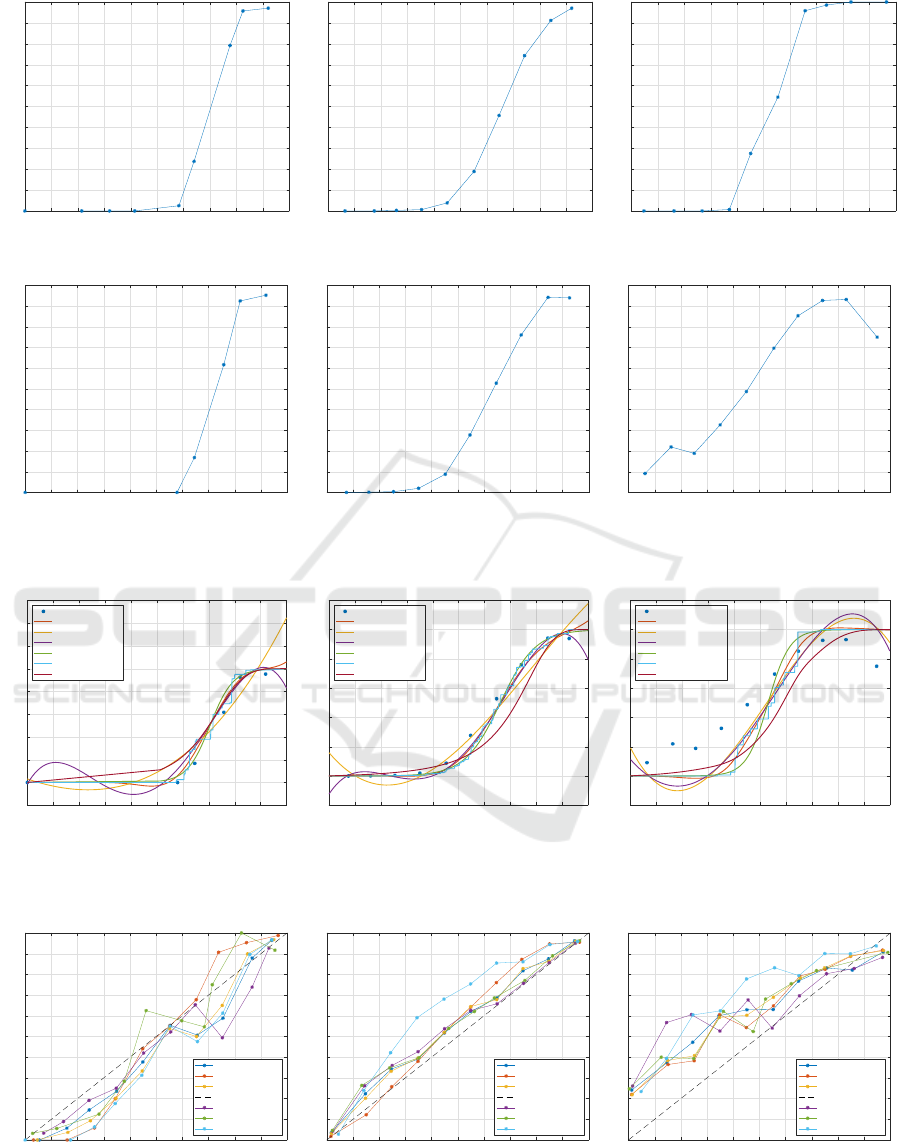
In Figure 1 we show the reliability diagrams for train-
ing and test set for the three datasets. The binning
has been performed with 10 uniformly-spaced bins, a
common strategy in related works. In all cases, the
trend is way far from the y = x diagonal line: a clear
sign that the classifier is not well-calibrated. By con-
sidering the ADULT dataset (training set) as an exam-
ple, it is possible to see that all points whose score is
less than 0.7 lie below the y = x line: this means that
all points with score (as returned by SVM) less than or
equal to 0.7 have probability to belong to class 1 way
inferior with respect to the score itself; similarly, for
all points with score greater than 0.7, the true proba-
bility to belong to class 1 is superior with respect to
the score assigned by the classifier.
Figure 2 shows the results in terms of fitted curves
over the reliability diagram on the test set for the three
considered datasets. Conversely, in Figure 3 we show
the reliability diagram after calibration. For ease of
comparison, in Tables 1 and 2 we show the two fig-
ure of merits (Brier score and Log-Loss score, respec-
tively) on both the training set and test set.
By considering the performances on test set, it
is possible to see that the three alternative methods
(Poly3, Poly4, NCS) have Brier score comparable to
state-of-the-art techniques (PS, IR, SC): Poly4 is the
best method for ABALONE, Poly3 and Poly4 equally
outperform other methods for ADULT and Poly3 is the
best method for PCN. In terms of Log-Loss, PS is the
best method for ABALONE and ADULT, whereas SC
is the best for PCN. Furthermore, the three alterna-
tive methods are featured by a lower computational
burden, being a plain curve fitting over the reliability
diagram.
4 CONCLUSIONS
In this paper we reviewed three state-of-the-art tech-
niques for calibrating a binary classifier in order to
return reliable probability estimates on the resulting
predictions. The three techniques (PS, IR and SC)
have been benchmarked on two well-known datasets
(ABALONE and ADULT) and an additional dataset
(PCN) against three lightweight methods (Poly3,
Poly4 and NSC), which basically perform a plain
curve fitting on the reliability diagram. Computa-
tional results show that the three methods are com-
parable in terms of Brier score and Log-Loss score
with respect to the three state-of-the-art approaches.
For these tests we used a SVM classifier due to its
uncalibrated behaviour and in order to stress the com-
parison amongst calibration techniques rather than
classification systems. Nonetheless, future research
endeavours will consider the application of such tech-
niques to different classification systems.
Indeed this study is part of a wider project concern-
ing the design and implementation of a modelling and
recognition system of faults and outages occurring
in the real-world power grid managed by “Azienda
Comunale Energia e Ambiente” (ACEA) company in
Rome, Italy. The recognition system, based on a one-
class classification approach as the main core of a
larger system (De Santis et al., 2015), has been devel-
oped within the “ACEA Smart Grids project”. A first
task consists in modelling and recognizing faults in
the power grid within a Decision Support System that
provides support for the commanding and dispatching
system, aiming at the implementation of Condition
Based Maintenance programs. Another very impor-
tant task consists in extracting from the learned fault
classification model useful information for program-
ming and control procedures, such as the estimation
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
492
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(a) ABALONE (training set)
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(b) ADULT (training set)
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(c) PCN (training set)
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(d) ABALONE (test set)
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(e) ADULT (test set)
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
(f) PCN (test set)
Figure 1: Reliability Diagrams.
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
-0.2
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
Fraction of positive instances (empirical probabilities)
Reliability Diagram
NCS
Poly3
Poly4
PS
IR
SC
(a) ABALONE
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
-0.2
0
0.2
0.4
0.6
0.8
1
1.2
Fraction of positive instances (empirical probabilities)
Reliability Diagram
NCS
Poly3
Poly4
PS
IR
SC
(b) ADULT
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
-0.2
0
0.2
0.4
0.6
0.8
1
1.2
Fraction of positive instances (empirical probabilities)
Reliability Diagram
NCS
Poly3
Poly4
PS
IR
SC
(c) PCN
Figure 2: Reliability Diagrams vs. fitted curves. For ABALONE we observe that for x ∈ (0,0.6) blue asterisks are missing,
meaning that there are no scores in such bins.
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
NCS
Poly3
Poly4
Perfect Calibration
PS
IR
SC
(a) ABALONE
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
NCS
Poly3
Poly4
Perfect Calibration
PS
IR
SC
(b) ADULT
0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1
Mean predicted value (score)
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Fraction of positive instances (empirical probabilities)
NCS
Poly3
Poly4
Perfect Calibration
PS
IR
SC
(c) PCN
Figure 3: Reliability Diagrams after Calibration.
Calibration Techniques for Binary Classification Problems: A Comparative Analysis
493

Table 1: Brier Score.
Method Abalone Adult PCN
Training Set Test Set Training Set Test Set Training Set Test Set
uncalibrated 0.1788 0.1911 0.2028 0.1754 0.1302 0.1814
PS 0.1140 0.1209 0.1057 0.1084 0.0488 0.1977
IR 0.1083 0.1215 0.1050 0.1081 0.0442 0.1934
SC 0.1189 0.1263 0.1143 0.1179 0.0767 0.2073
Poly3 0.1368 0.1355 0.1095 0.1069 0.0582 0.1822
Poly4 0.1172 0.1236 0.1059 0.1069 0.0579 0.1847
NCS 0.1142 0.1207 0.1057 0.1070 0.0493 0.1878
Table 2: Log-Loss Score.
Method Abalone Adult PCN
Training Set Test Set Training Set Test Set Training Set Test Set
uncalibrated 0.5293 0.550 0.5919 0.5306 0.4404 0.5508
PS 0.3711 0.3901 0.3301 0.3395 0.1716 0.8451
IR 0.3503 0.3943 0.3274 0.3453 0.1476 2.3263
SC 0.3938 0.4080 0.3573 0.3626 0.2867 0.6486
Poly3 0.4572 0.4586 0.3613 0.3807 0.2260 1.8316
Poly4 0.3865 0.4184 0.3660 0.4416 0.2245 1.7577
NCS 0.3865 0.3909 0.3406 0.3718 0.1843 2.0713
of the financial risk associated to a set of power grid
states and network resilience analysis. When dealing
with risk assessment and cost benefit analysis for net-
work expansion and maintenance planning, the avail-
ability of reliable probability estimates is of utmost
importance.
REFERENCES
Ayer, M., Brunk, H. D., Ewing, G. M., Reid, W. T., and
Silverman, E. (1955). An empirical distribution func-
tion for sampling with incomplete information. The
Annals of Mathematical Statistics, 26(4):641–647.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat,
T., Weissig, H., Shindyalov, I. N., and Bourne, P. E.
(2000). The protein data bank. Nucleic Acids Re-
search, 28(1):235–242.
Boser, B. E., Guyon, I., and Vapnik, V. (1992). A training
algorithm for optimal margin classifiers. In Proceed-
ings of the fifth annual workshop on Computational
learning theory, pages 144–152. ACM.
Brier, G. W. (1950). Verification of forecast expressed
in terms of probability. Monthly Weather Review,
78(1):1–3.
Butler, S. (2016). Algebraic aspects of the normalized
Laplacian, pages 295–315. Springer International
Publishing, Cham.
Cortes, C. and Vapnik, V. (1995). Support-vector networks.
Machine learning, 20(3):273–297.
Cristianini, N. and Shawe-Taylor, J. (2000). An Introduction
to Support Vector Machines and Other Kernel-based
Learning Methods. Cambridge University Press.
De Santis, E., Livi, L., Sadeghian, A., and Rizzi, A. (2015).
Modeling and recognition of smart grid faults by a
combined approach of dissimilarity learning and one-
class classification. Neurocomputing, 170:368 – 383.
De Santis, E., Martino, A., Rizzi, A., and Frattale Mascioli,
F. M. (2018a). Dissimilarity space representations and
automatic feature selection for protein function pre-
diction. In 2018 International Joint Conference on
Neural Networks (IJCNN), pages 1–8.
De Santis, E., Paschero, M., Rizzi, A., and Frattale Mas-
cioli, F. M. (2018b). Evolutionary optimization of
an affine model for vulnerability characterization in
smart grids. In 2018 International Joint Conference
on Neural Networks (IJCNN), pages 1–8.
DeGroot, M. H. and Fienberg, S. E. (1983). The com-
parison and evaluation of forecasters. Journal of the
Royal Statistical Society. Series D (The Statistician),
32(1/2):12–22.
Di Noia, A., Martino, A., Montanari, P., and Rizzi, A.
(2019). Supervised machine learning techniques and
genetic optimization for occupational diseases risk
prediction. Soft Computing.
Di Paola, L., De Ruvo, M., Paci, P., Santoni, D., and
Giuliani, A. (2012). Protein contact networks: an
emerging paradigm in chemistry. Chemical Reviews,
113(3):1598–1613.
Domingos, P. M. and Pazzani, M. J. (1996). Beyond inde-
pendence: Conditions for the optimality of the simple
bayesian classifier. In Proceedings of the Thirteenth
NCTA 2019 - 11th International Conference on Neural Computation Theory and Applications
494

International Conference on International Conference
on Machine Learning, ICML’96, pages 105–112, San
Francisco, USA. Morgan Kaufmann Publishers Inc.
Dua, D. and Graff, C. (2019). UCI machine learning repos-
itory. http://archive.ics.uci.edu/ml.
Freedman, D. and Diaconis, P. (1981). On the histogram
as a density estimator: L2 theory. Zeitschrift f
¨
ur
Wahrscheinlichkeitstheorie und Verwandte Gebiete,
57(4):453–476.
Hastie, T., Tibshirani, R., and Friedman, J. (2001). The
elements of statistical learning. Springer-Verlag, New
York, USA.
Jain, A. K., Murty, M. N., and Flynn, P. J. (1999). Data
clustering: a review. ACM computing surveys (CSUR),
31(3):264–323.
Li, Y. H., Xu, J. Y., Tao, L., Li, X. F., Li, S., Zeng, X.,
Chen, S. Y., Zhang, P., Qin, C., Zhang, C., Chen, Z.,
Zhu, F., and Chen, Y. Z. (2016). Svm-prot 2016: A
web-server for machine learning prediction of protein
functional families from sequence irrespective of sim-
ilarity. PLOS ONE, 11(8):1–14.
Lin, H.-T., Lin, C.-J., and Weng, R. C. (2007). A note
on platt’s probabilistic outputs for support vector ma-
chines. Machine Learning, 68(3):267–276.
Lucena, B. (2018). Spline-based probability calibration.
arXiv preprint arXiv:1809.07751.
Maiorino, E., Rizzi, A., Sadeghian, A., and Giuliani, A.
(2017). Spectral reconstruction of protein contact net-
works. Physica A: Statistical Mechanics and its Ap-
plications, 471:804 – 817.
Martino, A., Giuliani, A., and Rizzi, A. (2018a). Gran-
ular computing techniques for bioinformatics pat-
tern recognition problems in non-metric spaces. In
Pedrycz, W. and Chen, S.-M., editors, Computational
Intelligence for Pattern Recognition, pages 53–81.
Springer International Publishing, Cham.
Martino, A., Maiorino, E., Giuliani, A., Giampieri, M., and
Rizzi, A. (2017a). Supervised approaches for function
prediction of proteins contact networks from topolog-
ical structure information. In Sharma, P. and Bianchi,
F. M., editors, Image Analysis, pages 285–296, Cham.
Springer International Publishing.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2017b).
Efficient approaches for solving the large-scale k-
medoids problem. In Proceedings of the 9th Inter-
national Joint Conference on Computational Intelli-
gence - Volume 1: IJCCI,, pages 338–347. INSTICC,
SciTePress.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2018b).
Distance matrix pre-caching and distributed computa-
tion of internal validation indices in k-medoids clus-
tering. In 2018 International Joint Conference on
Neural Networks (IJCNN), pages 1–8.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M. (2018c).
Supervised approaches for protein function prediction
by topological data analysis. In 2018 International
Joint Conference on Neural Networks (IJCNN), pages
1–8.
Martino, A., Rizzi, A., and Frattale Mascioli, F. M.
(2019). Efficient approaches for solving the large-
scale k-medoids problem: Towards structured data.
In Sabourin, C., Merelo, J. J., Madani, K., and War-
wick, K., editors, Computational Intelligence: 9th In-
ternational Joint Conference, IJCCI 2017 Funchal-
Madeira, Portugal, November 1-3, 2017 Revised Se-
lected Papers, pages 199–219. Springer International
Publishing, Cham.
Minneci, F., Piovesan, D., Cozzetto, D., and Jones, D. T.
(2013). Ffpred 2.0: Improved homology-independent
prediction of gene ontology terms for eukaryotic pro-
tein sequences. PLOS ONE, 8(5):1–10.
Murphy, A. H. and Winkler, R. L. (1977). Reliability of sub-
jective probability forecasts of precipitation and tem-
perature. Journal of the Royal Statistical Society. Se-
ries C (Applied Statistics), 26(1):41–47.
Naeini, M. P., Cooper, G. F., and Hauskrecht, M. (2015).
Obtaining well calibrated probabilities using bayesian
binning. In Proceedings of the Twenty-Ninth AAAI
Conference on Artificial Intelligence, AAAI’15, pages
2901–2907. AAAI Press.
Niculescu-Mizil, A. and Caruana, R. (2005). Predicting
good probabilities with supervised learning. In Pro-
ceedings of the 22nd international conference on Ma-
chine learning, pages 625–632. ACM.
Parzen, E. (1962). On estimation of a probability den-
sity function and mode. The Annals of Mathematical
Statistics, 33(3):1065–1076.
Platt, J. (2000). Probabilities for sv machines. In Smola,
A. J., Bartlett, P., Sch
¨
olkopf, B., and Schuurmans, D.,
editors, Advances in large margin classifiers, pages
61–74. MIT Press, Cambridge, MA, USA.
Sch
¨
olkopf, B. and Smola, A. J. (2002). Learning with ker-
nels: support vector machines, regularization, opti-
mization, and beyond. MIT Press.
Scott, D. W. (1979). On optimal and data-based histograms.
Biometrika, 66(3):605–610.
Sturges, H. A. (1926). The choice of a class inter-
val. Journal of the American Statistical Association,
21(153):65–66.
The UniProt Consortium (2017). Uniprot: the univer-
sal protein knowledgebase. Nucleic Acids Research,
45(D1):D158–D169.
Wahba, G. (1990). Spline models for observational data,
volume 59. Siam.
Webb, E. C. (1992). Enzyme nomenclature 1992. Recom-
mendations of the Nomenclature Committee of the In-
ternational Union of Biochemistry and Molecular Bi-
ology on the Nomenclature and Classification of En-
zymes. Academic Press, 6 edition.
Zadrozny, B. and Elkan, C. (2001). Obtaining calibrated
probability estimates from decision trees and naive
bayesian classifiers. In Proceedings of the Eigh-
teenth International Conference on Machine Learn-
ing, ICML ’01, pages 609–616, San Francisco, CA,
USA. Morgan Kaufmann Publishers Inc.
Zadrozny, B. and Elkan, C. (2002). Transforming classifier
scores into accurate multiclass probability estimates.
In Proceedings of the eighth ACM SIGKDD interna-
tional conference on Knowledge discovery and data
mining, pages 694–699. ACM.
Calibration Techniques for Binary Classification Problems: A Comparative Analysis
495
