
Multimodal Ranked Search over Integrated Repository of Radiology
Data Sources
Priya Deshpande
a
, Alexander Rasin, Fang Cao, Sriram Yarlagadda, Eli Brown,
Jacob Furst and Daniela S. Raicu
College of Computing and Digital Media, DePaul University, Chicago, U.S.A.
Keywords:
Information Retrieval, Relevance Ranking, Radiology Teaching Files Database, Medical Ontology,
Content based Image Retrieval.
Abstract:
Radiology teaching files serve as a reference in the diagnosis process and as a learning resource for radiology
residents. Many public teaching file data sources are available online and private in-house repositories are
maintained in most hospitals. However, the native interfaces for querying public repositories have limited
capabilities. The Integrated Radiology Image Search (IRIS) Engine was designed to combine public data
sources and in-house teaching files into a single resource. In this paper, we present and evaluate ranking
strategies that prioritize the most relevant teaching files for a query. We quantify query context through a
weighted text-based search and with ontology integration. We also incorporate an image-based search that
allows finding visually similar teaching files. Finally, we augment text-based search results with image-based
search – a hybrid approach that further improves search result relevance. We demonstrate that this novel
approach to searching radiology data produces promising results by evaluating it with an expert panel of
reviewers and by comparing our search performance against other publicly available search engines.
1 INTRODUCTION
Vast amounts of image and clinical report data such
as Electronic Health Records (EHRs), pathology re-
ports, and teaching files are generated in the health-
care domain. Teaching files are used by radiologists,
residents, and medical students (Dashevsky et al.,
2015) as a reference resource. A typical teaching file
contains text data categories such as (patient) history,
findings, diagnosis, discussion, references, and case
images. To find the relevant teaching files, radiol-
ogists need a domain-aware search engine that inte-
grates diverse data sources. To find the most relevant
material for a particular case, a search tool should
support a hybrid text and image (multimodal) search.
(Simpson et al., 2014) argued that the integration
of text-based and image-based search could improve
medical data retrieval.
Several studies highlighted the need to integrate
clinical reports and images into a database with ad-
vanced search capabilities. (Gutmark et al., 2007)
argued for building a system that reduces errors in
radiological images interpretation using teaching file
a
https://orcid.org/0000-0002-2631-5751
databases. (Talanow, 2009) described how critical ref-
erence radiological images are for diagnosis, teaching
needs, and research. (Pinho et al., 2017) investigated
the difficulties inherent to the content-based image re-
trieval (CBIR) in Picture Archiving and Communi-
cation Systems (PACS). (Russell-Rose and Chamber-
lain, 2017) performed a study of healthcare informa-
tion professionals needs, goals, and requirements for
information retrieval systems. (Li et al., 2018) pro-
posed a hybrid retrieval-generation system, where hu-
man knowledge and traditional retrieval systems are
used to generate reports. (Ling et al., 2014) designed
GEMINI, an integrative healthcare analytics system
and investigated integration of heterogeneous data.
The study concluded the healthcare needs are not met
by the current search engines. We surveyed the pub-
licly available search tools for teaching file data (see
Section 2.2.2) and found that they do not rank search
results based on query relevance. Engines that focus
on medical articles perform better; however, research
articles are not typically used for diagnosis. We evalu-
ate public search engine query results in Section 4.1.3.
Our approach is presented and evaluated in the
context of the IRIS (Deshpande et al., 2017) and
(Deshpande et al., 2018b), using a major public
372
Deshpande, P., Rasin, A., Cao, F., Yarlagadda, S., Brown, E., Furst, J. and Raicu, D.
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources.
DOI: 10.5220/0008166603720383
In Proceedings of the 11th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2019), pages 372-383
ISBN: 978-989-758-382-7
Copyright
c
2019 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

teaching file data source, Medical Imaging Resource
Community (MIRC
1
) with two major medical on-
tologies Radiology Lexicon (RadLex
2
) and System-
atized Nomenclature of Medicine Clinical Terms
(SNOMED CT
3
). Based on feedback from radiol-
ogy domain experts, we also augmented our retrieval
mechanism to apply different weights to categories of
text in teaching files. In cases where two or more
teaching files are assigned the same relevance score,
we apply our co-occurrence algorithm to re-rank the
results from the tie point. For example, consider a
query “progeria”: “cardiomegaly” often occurs in pa-
tients with “progeria”. Thus, we increase the rele-
vance of teaching files that include “cardiomegaly”.
Our contributions presented in this paper are:
• Weighted text based search with integrated medi-
cal ontologies to provide search relevance ranking
• Image based search
• Hybrid search that augments text results based on
an image search
• An evaluation of IRIS and other search engines
In the rest of the paper Section 2 presents related
work; Section 3 discusses our system implementation
and design choices; Section 4 analyses the outcomes
of our system evaluation. Finally, Section 5 presents
our key conclusions.
2 BACKGROUND AND RELATED
WORK
Our system has been influenced by numerous public
search engines. In this section, we first discuss the
need for a full featured radiology teaching file search
engine. We then review existing public repository
sources and medical ontologies.
2.1 Biomedical Search Engines
(Dos-Santos and Fujino, 2012) discussed the need for
the integration of profiles published by Integrating the
Healthcare Enterprise (IHE) and providing the access
to cases through IHE. (Pinho et al., 2017) proposed an
extensible platform for multimodal medical image re-
trieval, integrated in an open-source PACS software.
In their platform, image queries rely on a query-by-
example pattern, supporting only the images from a
pre-indexed dataset. (Simpson et al., 2014) proposed
1
http://mirc.rsna.org/query
2
http://www.radlex.org/
3
https://www.nlm.nih.gov/healthit/snomedct
a multimodal image retrieval system that retrieves
biomedical articles used in Open-i
4
(evaluated in Sec-
tion 4.1.3). (Hwang et al., 2016) shows how the use
of positron emission tomography-computed tomogra-
phy increased the need to retrieve relevant medical
images to assist image interpretation. Furthermore,
(Kansagra et al., 2016) presented the idea of a global
database that integrates multiple data sources for a
more accurate diagnoses.
Although most radiology systems have relied on
text queries, Content-based Image Retrieval (CBIR)
has also been used in the past. The survey by (Akg
¨
ul
et al., 2011) that outlines recent work concluded that
the lack of a common image database and differences
in the application domains makes comparative analy-
sis of CBIR approaches difficult. (M
¨
uller et al., 2001)
defined a set of evaluation guidelines developed for
CBIR systems, which we considered for our CBIR
evaluation.
Both literature survey and radiologists we con-
sulted confirmed the need for a domain-tailored teach-
ing file search engine. As we show in this paper,
public radiology teaching file search engines do not
provide relevance ranking. Our contributions pre-
sented in this paper are domain-specific, presenting
a use case for a reference search source in radi-
ology diagnostics. While we rely on standard co-
occurrence information retrieval techniques, we de-
signed a weighted ontology co-occurrence algorithm
and a hybrid multimodal search algorithm specifically
for the identified need within the radiology domain.
Our data integration work is presented in (Deshpande
et al., 2018a) and (Deshpande et al., 2019b). IRIS
served as a prototype for our biomedical data integra-
tion and indexing system (Deshpande et al., 2019a).
We presented IRIS at a major medical conference and
received feedback from doctors indicating that this
work would be useful for domain practitioners.
2.2 Data Sources and Search Engines
In this section, we compare available data repositories
and medical search engines (summarized in Table 1).
2.2.1 Teaching File Data Sources
RSNA MIRC. Radiology Society of North Amer-
ica Medical Imaging Resource Community is a large
repository with more than 2,500 teaching files and
more than 12,000 images as well as external refer-
ences (journal articles). Text search is done verbatim
with no processing to interpret the user’s query (e.g.,
considering term synonyms or negation).
4
https://openi.nlm.nih.gov/
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
373
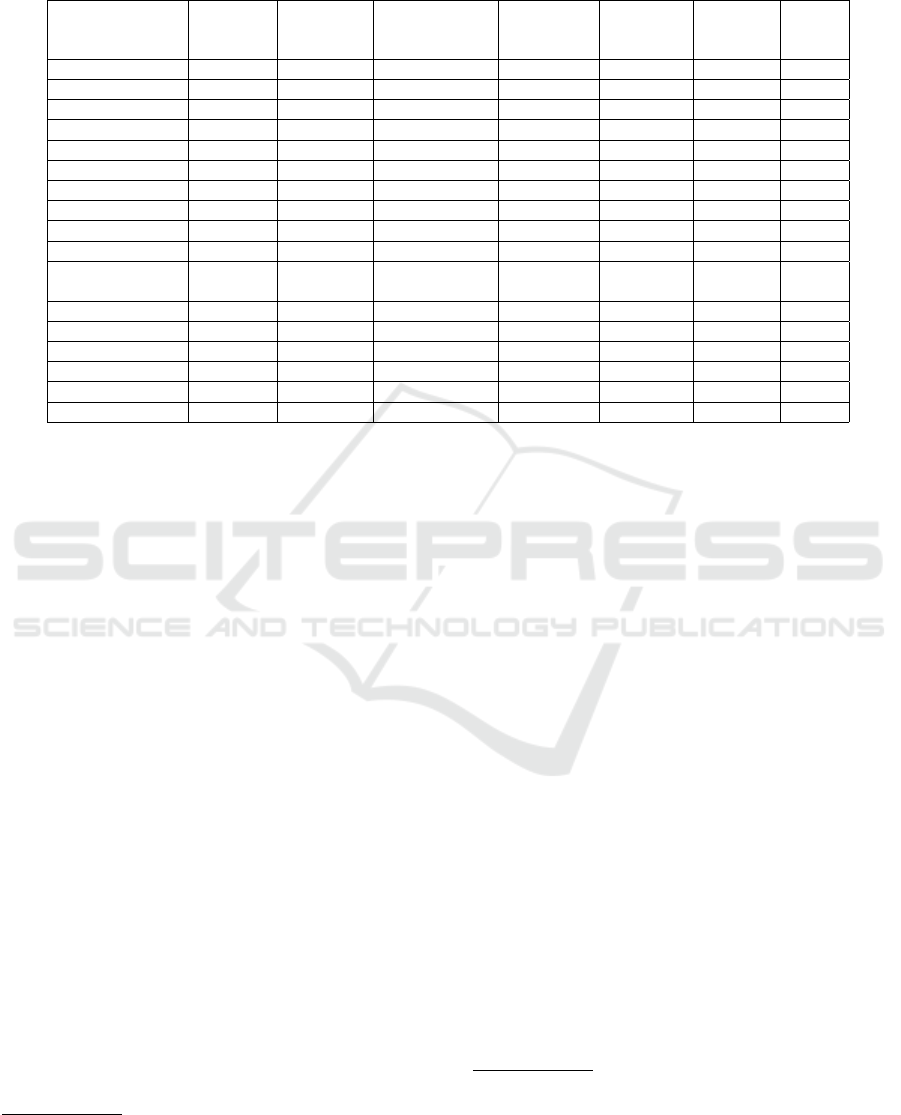
Table 1: A comparative study of available data sources and search engines NLP capabilities. Relationships between terms:
Hierarchical relation between terms (is a/has a), Spelling Error Correction: Prompt user with correct spelling, Relevance
Rank: Offers an explicit relevance ranking feature.
Search
Engine
Keyword
search
Synonyms
Relationships
between terms
Spelling
Error
Correction
Relevance
Rank
publicly
available
Image
search
RadTF YES YES YES NO NO NO NO
GoldMiner YES NO NO YES NO YES NO
Yottalook YES YES NO YES YES YES NO
Google YES NO NO YES YES YES YES
MIRC YES NO NO NO NO YES NO
MyPacs YES NO NO NO NO YES NO
CTisus YES NO NO YES NO YES NO
Casimage NO NO NO NO NO NO NO
RadICS NO NO NO NO NO NO NO
BIMM YES NO NO NO NO NO NO
Radiology
Teacher
YES NO NO NO NO YES NO
Medscape YES NO NO YES NO YES NO
ImageCLEFmed NO NO NO NO NO NO NO
Khresmoi YES YES NO NO NO YES NO
Openi YES YES NO YES YES NO YES
EURORAD YES NO NO NO NO YES NO
IRIS YES YES NO NO YES YES YES
Mypacs.net (Weinberger et al., 2002). A publicly
available teaching file resource, where radiologists
can upload new teaching files. More than 35,000
cases are available with 200,000 images. User can
search records based on anatomy, pathology, modal-
ity, age, gender, etc. However, the built-in search en-
gine has no consideration of synonyms, negation, or
context. No image-based search is available.
EURORAD.
5
European Society of Radiology is a
peer-reviewed educational tool based on teaching
cases. There are 7,000+ teaching cases – similarly
to other teaching file sources there is no support for
negation, synonyms, or image-based search.
A sample teaching file from both MIRC and My-
Pacs is shown in Figure 1, illustrating the inherent het-
erogeneity of such data sources.
2.2.2 Medical Search Engines
Yottalook.
6
A radiologist-targeted search engine
(“powered by Google Custom Search”). Searches a
variety of different sources such as radiopaedia.org,
American Journal of Radiology, University of Michi-
gan Medical School, and MyPacs.
Khresmoi.
7
is a medical informatics and retrieval
system that searches online biomedical information
and documents. Typical search result is a discussion
forum about a particular diagnosis or disease.
5
http://www.eurorad.org/
6
www.yottalook.com
7
http://everyone.khresmoi.eu/hon-search/
However, there are no teaching file cases in the
database and the search engine supports only ba-
sic query string matching with no NLP support.
MedGIFT.
8
projects are developed to advance the
field of medical visual information retrieval. All
tools are open sourced – however, these tools are
not a complete search engine. One of the projects,
ImageCLEF
9
, is the cross-language image retrieval
track providing benchmarks for CBIR systems.
NovaMedSearch.
10
is a medical search engine that
supports multimodal queries to find articles on the
PubMed Central in Clinical Decision Support scenar-
ios. There are no teaching files in this repository and
only a basic string matching search is supported.
Open-i. Open Access Biomedical Image Search
Engine of the National Library of Medicine enables
search and retrieval of abstracts and images (e.g.,
charts, graphs, clinical images) from the open source
literature and biomedical image collections. Search-
ing may be done using text queries as well as images.
CTisus.
11
A repository with more than 250,000
radiological images, quizzes and CT protocols are
available. However, their search engine does support
teaching file search or image based search.
Medscape.
12
A data source for latest medical news
and information about drugs and diseases. No
image-based search engine is available.
8
http://medgift.hevs.ch/wordpress/demos/
9
http://www.imageclef.org/FAQ
10
https://medical.novasearch.org/
11
http://www.ctisus.com/
12
http://www.medscape.com
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
374
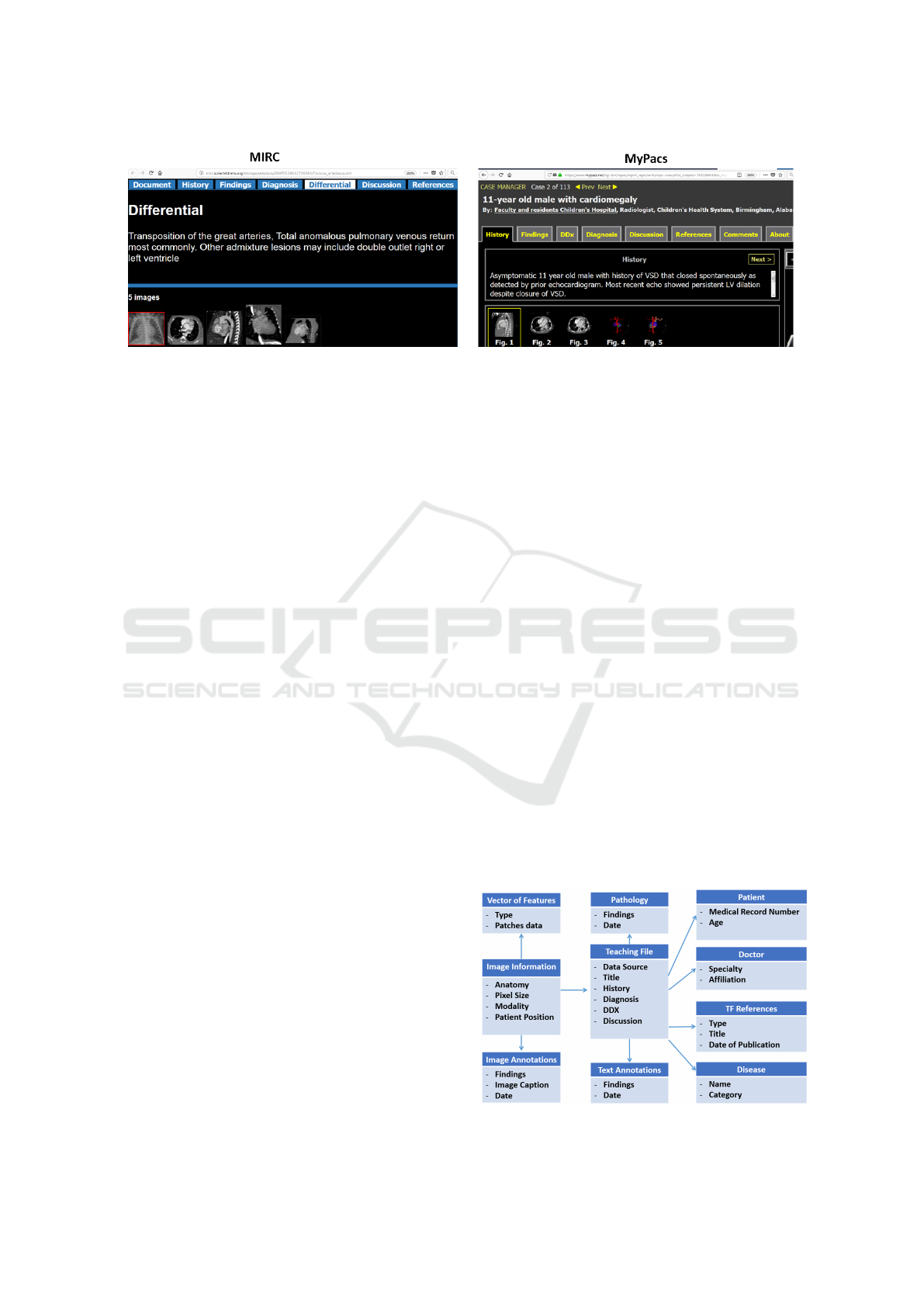
Figure 1: Sample teaching file from MIRC and MyPacs.
Casimage database with the IRMA framework
(Thies et al., 2004) An integration of a multime-
dia teaching and reference database in a Picture
Archiving and Communication Systems (PACS)
environment. This source is not publicly available.
RADTF. (Do et al., 2010) A teaching file repository
that uses NLP to ingest radiology reports. Search en-
gine uses RadLex anatomy concept terms, stemming,
ranking of results based on negation or uncertainty
expressions. RADTF is not publicly available – the
link provided in (Do et al., 2010) is no longer active.
Server-based Digital Teaching File System
RADICS. (Kamauu et al., 2006) The RadICS server
could handle CT, MR, computed radiography, and
digital radiography images adhering to the Digital
Imaging and Communications in Medicine (DICOM)
format. The engine is not publicly available.
Biomedical Image Metadata Manager (BIMM)
(Korenblum et al., 2011). provides retrieval of
similar images using semantic features. Not available
– the link in (Korenblum et al., 2011) is inactive.
The caBIG
TM
Annotation and Image Markup
Project (Channin et al., 2010). developed a mech-
anism for modeling, capture and serializing image
annotation, readable by both human and machine.
The link in the paper is no longer available.
Radiology Teacher (Talanow, 2009). A web-based
teaching file development system. Allows authors
to create, edit, and delete cases and images with
annotations and offers educational quizzes. Contains
only about 300 cases; no NLP support is available.
2.3 Medical Ontologies
To build an effective search engine with medical do-
main knowledge, we integrated two medical ontolo-
gies (RadLex and SNOMED CT) into IRIS.
RadLex. is an ontological system that provides a
comprehensive lexicon vocabulary for radiologists.
Developed by Radiology Society of North America
(RSNA), RadLex defines over 45,000 unique terms.
The Systematized Nomenclature of Medicine Clin-
ical Terms (SNOMED CT). ontology provides a
standardized, multilingual vocabulary of clinical ter-
minology that is used by physicians and healthcare
providers for the electronic exchange of clinical infor-
mation. The SNOMED CT ontology follows the Na-
tional Library of Medicine (NLM) Unified Medical
Language System format; it has a hierarchical struc-
ture that describes morphological term connections.
3 METHODOLOGY
3.1 IRIS Architecture and Query Flow
We designed a logical schema (a subset of the schema
is shown in Figure 2, other attributes and entities were
omitted) and extracted and loaded data from publicly
available radiology teaching files data repositories.
As part of our data loading, we performed data clean-
ing (e.g., removal of unnecessary stop-words, invalid
dates). The base entry in the center of the schema is
a teaching file record linked with a collection of im-
age entries, since teaching files are bundled with im-
ages (e.g., MRI, X-ray). The vector of features table
contains extracted patches – a user-defined data type
Figure 2: Logical Schema used by IRIS.
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
375
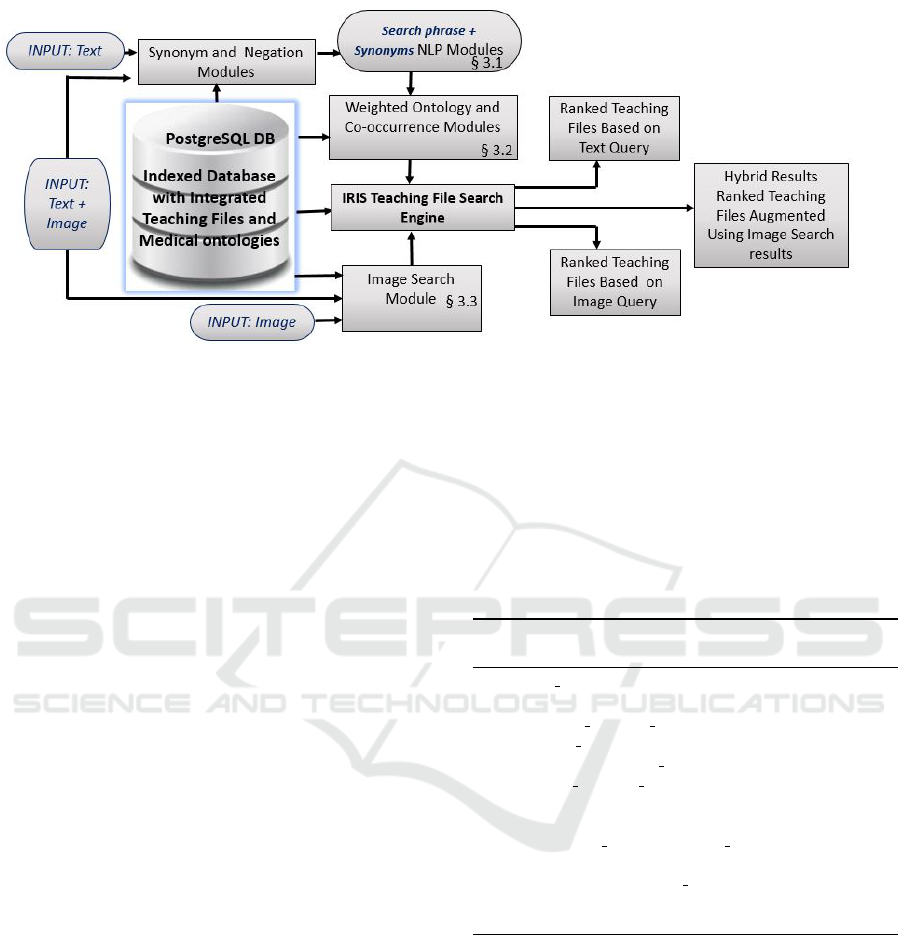
Figure 3: IRIS Query Execution Flow.
describing a region of interest marked by radiologists.
For text-based search, we generate a matrix based
on term frequency, and term co-occurrence (see Sec-
tion 3.2.2) from the teaching file terms. To expedite
matrix creation, we pre-computed a term table that in-
dexes both single-word term and multi-word ontology
terms as they appear in teaching files. To keep the in-
tegrated datasets up-to-date we run a periodic refresh
based on “date of modification” in source reposito-
ries, propagating the changes into the database.
Figure 3 shows the execution flow in IRIS en-
gine. IRIS uses a Natural Language Processing
(NLP) module that performs text query expansion
using integrated ontologies and substitutes negation
with antonyms. Results are ranked using an ontology
based weighted co-occurrence matrix. To perform an
image search, IRIS returns teaching files that contain
the matched images. When both text and image query
inputs are specified, text search results are filtered and
re-organized using the outcome of the image search.
3.2 Text-based Search
In this section we discuss ranking features of our
search algorithm: 1) Adding weights to teaching files
categories and 2) Generating a co-occurrence matrix
to break ties. Text search is evaluated in Section 4.1.
3.2.1 Weighted Category and Ontology
Integration with NLP Support
Based on a literature survey and feedback from radi-
ologists, we chose to apply the highest relative weight
(4) to terms which belong to title, findings, and diag-
nosis categories within the teaching file. A weight
of 3 is assigned to history and differential diagnosis
categories. Finally, the lowest relative weight was as-
signed to discussion category (weight of 2) and ref-
erences category (weight of 1). We furthermore dif-
ferentiated the medical ontology from non-ontology
terms by introducing an additional weight of 1. Al-
gorithm 1 outlines the adding of weight to differ-
ent category terms when generating a teaching file
term vector. A teaching file term vector contains all
unique terms and the associated term frequency of
those terms based on the assigned weights.
Algorithm 1: Generating the matrix with weighted category
and ontology terms.
1: allterm vector ← all teaching file (TF) terms
2: Matrix = []
3: for t f in all teaching f iles do
4: Matrix row = []
5: for term in allterm vector do
6: all category count ← Term frequency by
TF category
Σ
categories
Count
category
×Weight
category
7: Matrix row.append(term score)
8: end for
9: Matrix.append(Matrix row)
10: end for
11: return Matrix
As in previous work, IRIS incorporates synonym
and negation interpretation, allowing users to search
for ”with X”, or ”without X”, and similar constructs.
Our search engine automatically performs query ex-
pansion using integrated ontologies. For example, if
a user searches for “breast cancer”, we augment the
search with additional terms such as “malignant tu-
mor of breast” and “carcinoma of breast”.
3.2.2 Weighted Co-occurrence Matrix
A term-weighted search may produce a tied score.
This is particularly common when there are relatively
few matches (and thus not a lot of different teaching
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
376

files to differentiate). We therefore extended our algo-
rithm by introducing a weighted co-occurrence ma-
trix that is used to break ties and to find additional
results based on term co-occurrence. To build a co-
occurrence matrix, teaching file data are mapped into
a matrix T called a term frequency matrix, where T
i j
is the frequency of term j in the teaching file i with
j=1,. . . , m; i=1,. . . ,n, and m and n are the number of
terms and teaching files, respectively. A query string
q is encoded the same way as a teaching file, i.e., as a
column vector with an element corresponding to each
term from the document corpus. With this represen-
tation, a query search can be thought of as a simple
linear algebra operation, specifically the product be-
tween T and q (Equation 1). The query search result r
is represented as a vector in which each element pro-
vides a score for how well the teaching file document
matches the query string q.
r1
r2
.
.
.
rn
=
T
11
T
12
. . . T
1m
T
21
T
22
. . . T
2m
.
.
.
.
.
.
.
.
.
.
.
.
T
n1
T
n2
. . . T
nm
•
q1
q2
.
.
.
qm
(1)
For all teaching files, we denote by C
term
the co-
occurrence matrix of all m terms and C
(RS raw)
(RS:
RadLex and SNOMED CT) the co-occurrence ma-
trix of all k RadLex and SNOMED CT terms. In the
eventuality that the two co-occurrence matrices have
different sizes (as in the case of MIRC where only
18% of the terms belong to RadLex and 36% terms
belongs to SNOMED CT), the C
(RS
raw)
matrix is ex-
tended with the identity matrix I (Equation 2):
C
RS
=
C
RS raw
0
0 I
(2)
The overall co-occurrence matrix C is then defined
as the dot product of C
term
and C
RS
in order to assign
more weight to the co-occurrence of the RadLex and
SNOMED CT ontology terms (Equation 3):
C
11
C
12
. . . C
1m
C
21
C
22
. . . C
2m
.
.
.
.
.
.
.
.
.
.
.
.
C
m1
C
m2
. . . C
mm
=
C
term
11
C
term
12
. . . C
term
1m
C
term
21
C
term
22
. . . C
term
2m
.
.
.
.
.
.
.
.
.
.
.
.
C
term
m1
C
term
n2
. . . C
term
mm
•
C
RS raw
0
0 I
(3)
The co-occurrence matrix C is then applied to the
formulation of Equation 1, effectively including co-
occurrence of terms (i.e. context) and medical ontolo-
gies in the relevance calculation as shown in Equa-
tion 4. The transformation used in Equation 4 is pre-
computed offline to improve query response time.
When multiple teaching files receive the same rel-
evance score, IRIS invokes the co-occurrence algo-
rithm to break the tie. Search results up to the tie are
kept, while the results from the tie and below are re-
ranked using the co-occurrence matrix computation
of the relevance vector r. We kept search results be-
fore tie as it is because our search engine is domain
specific and appearance of search term in particular
category (e.g., diagnosis, findings) matters a lot. If we
apply only co-occurrence top results and less relevant
than we got using our weighted category algorithm.
r1
r2
.
.
.
rn
=
T
11
T
12
. . . T
1m
T
21
T
22
. . . T
2m
.
.
.
.
.
.
.
.
.
.
.
.
T
n1
T
n2
. . . T
nm
•
C
11
C
12
. . . C
1m
C
21
C
22
. . . C
2m
.
.
.
.
.
.
.
.
.
.
.
.
C
m1
C
m2
. . . C
mm
•
q1
q2
.
.
.
qm
(4)
3.3 Content based Image Retrieval
CBIR system retrieves images similar to an image
query. Typically, only the pixels of the image are used
for that purpose. Images are retrieved in the follow-
ing manner: an image feature extractor is used to ex-
tract latent features from the pixel array of all images
(both query and database images), then the features
of the query image can be compared to those of the
database images using a similarity measure. We con-
structed an image feature extractor using a convolu-
tional autoencoder. Convolutional autoencoders con-
sist of two main components - an encoder and a de-
coder. An encoder processes the original image into
an encoded image and decoder attempts to recreate
the original image from the encoded image. Since the
same autoencoder needs to work on a broad category
of images, the expectation is that the autoencoder will
likely capture the latent features that easily differenti-
ate one type of image from another.
Our convolutional autoencoder was built to accept
an image input of size 256 x 256 x 1 pixels. We
trained this network using 12,052 images obtained
from MIRC database. Prior to us inputting an image
into the network, we have first preprocessed it. Each
input image is converted to grayscale, its pixel inten-
sities are normalized to values between 0 and 255, and
resized to a size of 256 x 256. We used a trained au-
toencoder to encode images in the database after they
were preprocessed in a similar manner to the training
images. Each encoded image is then flattened into a
representative vector of length 8192. This vector is
used as the extracted features in the image retrieval
process. The search query feature vector is compared
to the feature vectors of the images from the database
through a (1-cosine) distance function and the most
similar images are retrieved, ordered by the distance
value. A (1-cosine) distance threshold of 0.12 is used
to filter what we consider false matches – any image
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
377

that is farther than 0.12 from the query image is re-
moved from consideration. This threshold is based on
the overall histogram distance distribution (top 10%
results) for all image vectors; a (1-cosine) distance
that is further than the nearest 10% of images is not
considered to be similar for our purposes.
3.4 Hybrid Search
In order to generate more relevant results, we im-
plemented a hybrid search algorithm that augments
text search with an image search. When a user is-
sues a text-and-image query, IRIS performs data fu-
sion by initially retrieving text search results, then
image search results, and then re-ranking text search
results by promoting cases that occurred in the im-
age search. Text based search, particularly when us-
ing co-occurrence matrix and ontologies, often results
in many teaching file matches. Moreover, text rele-
vance score does not consider the images in the teach-
ing file. IRIS hybrid text and image search algorithm
finds teaching files that explicitly mention the relevant
search terms (including ontology based synonyms);
image based search further prioritizes teaching files
that also have images similar to the query image.
The following example illustrates how our hybrid
search algorithm works (TC = Teaching Case) :
Text result ranking: TC5, TC7, TC2, TC9, TC11
Image search ranking: TC9, TC2, TC6
Hybrid search ranking: TC9, TC2, TC5, TC7, TC11
In hybrid search, TC9 and TC2 have increased pri-
ority due to appearing in both text and image searches,
TC6 is ignored because it did not match the text
search. TC5, TC7, and TC11 are kept but effectively
demoted because they only appear in the text search.
3.5 Evaluation Methodology
In this section we discuss the methodology for eval-
uation of IRIS search results. We used a combina-
tion of queries received from radiologists at a major
teaching hospital and other queries chosen from an
extensive literature survey (see Figure 4). We have
initially used 28 text queries, out of which we picked
a subset of 11 queries (listed in Table 2) to perform
an in depth evaluation. To choose these 11 queries,
we favored certain criteria such as: (1) queries where
the differential diagnosis and discussion categories
carry the same score for the query term (tie between
these two category cases – e.g., case with query term
“cardiomegaly” occurs in discussion category 3 times
and in diagnosis category 2 times). (2) queries with
relatively few results (to see if using co-occurrence
discovers additional results), (3) query terms which
Table 2: IRIS evaluation queries.
ID Query
Q1 Cardiomegaly
Q2 ACL tear
Q3 Annular pancreas
Q4 Pseudocoxalgia
Q5 Varicocele
Q6 Angiosarcoma
Q7 Tracheal dilation
Q8 Appendicitis
Q9 Bronchus intermedius
Q10 Cystitis glandularis
Q11 No cardiomegaly
are sufficiently precise for evaluation (e.g., “study” or
“toxic” are too general for a meaningful evaluation)
Our hybrid search evaluation with four experts
(with expertise in information retrieval but without
medical training) used a subset of 5 queries: “car-
diomegaly”, “no cardiomegaly”, “acl tear”, “tracheal
dilation”, and “angiosarcoma”. We used five queries
because it was not possible to collect a full evaluation
of 11 queries from a group of experts. We chose five
queries such that query results were relatively easy to
interpret by evaluators with no medical training.
Our search evaluation was based on a coding
standards document that included all relevant defi-
nitions (e.g,. medical term synonyms) and pertinent
information about the diseases (for evaluators with no
medical training). We defined five categories to score
text search results: “not relevant” = 0 (query term
and synonyms do not appear anywhere in the results),
“relevant” = 0.5 (term or synonyms appear in any
category of a teaching file), “more relevant” = 1 (if
term or synonyms appear in the discussion category),
“very relevant” = 1.5 (if term or synonyms appears
in history or ddx category), and “most relevant” =
2 (if term or synonyms appears in title, findings, or
diagnosis categories).
Text-based Search Evaluation. Our text-based
search ranking algorithm was evaluated by con-
sidering different combinations of search features.
We used weighted and unweighted category terms
(from teaching file categories) and weighted ontol-
ogy terms to generate co-occurrence matrix com-
bined with three possible search mechanisms (TF,
TF-IDF and co-occurrence) producing six possible
combinations shown in Table 3. The resulting six
categories were C
ut f
: unweighted term frequency,
C
ut f id f
: unweighted term frequency inverse document
frequency, C
uc
: unweighted co-occurrence, C
wt f
:
weighted term-frequency, C
wt f id f
: weighted term fre-
quency inverse document frequency, C
wc
: weighted
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
378

Figure 4: The outline of query evaluation of IRIS.
co-occurrence. One of the reasons for considering six
different categories was to compare TF-IDF which is
commonly used in search engines to TF (to verify that
TF-IDF is not needed in our domain). For text search
evaluation we used Normalized Discounted Cumula-
tive Gain (NDCG)
13
to measure the quality of search
result ranking. NDCG is based on Discounted cu-
mulative gain (DCG) – ”DCG measures the gain of
a document based on its position in the result list”.
NDCG is the normalized version of the DCG metric.
Cumulative Gain (CG) is the sum of the relevance
values of all results, as shown in Equation 5; rel
i
is
the relevance of the result at position i and p is a rank
position of the result. The definition of CG is included
here as background for the definition of DCG. We first
computed DCG (Equation 6) for top 10 search results.
CGp = Σ
p
i=1
rel
i
(5)
DCGp = Σ
p
i=1
2
rel
i
− 1
log
2
(i + 1)
(6)
To normalize the score, we computed ideal DCG
(IDCG), shown in Equation 7. IDCG first sorts all
known results by their relevance. |REL| represents
the ideally ordered list up to position p. We then com-
puted normalized DCG score based on Equation 8.
IDCGp = Σ
|REL|
i=1
2
rel
i
− 1
log
2
(i + 1)
(7)
nDCGp =
DCGp
IDCGp
(8)
Using this metric, we compared IRIS text-based
retrieval results with MIRC (which has no image-
based search). Our dataset and MIRC dataset contains
same teaching cases, making our evaluation particu-
larly consistent. We further compared IRIS to other
medical teaching file search engines.
Image-based Search Evaluation. There are no read-
ily available benchmarks against which we could
compare IRIS image-based results. We cannot com-
pare IRIS results with ImageCLEF (Tsikrika et al.,
2011) or TREC
14
as the data is completely differ-
13
https://en.wikipedia.org/wiki/Discounted cumulative
gain
14
http://www.trec-cds.org/2017.html
Table 3: Text-based search category abbreviations.
C
ut f
C
ut f id f
C
uc
C
wt f
C
wt f id f
C
wc
Weighted NO NO NO YES YES YES
Term Freq. YES NO YES YES NO YES
Term Freq. -
Inverse Doc.
Frequency
NO YES NO NO YES NO
Cooccurrence NO NO YES NO NO YES
ent. Therefore, we compared image-based results
with Google, and Open-i both of which provide im-
age search capabilities. Google is also a poor bench-
mark because it is a general purpose search engine,
while IRIS is a radiology domain-specific search en-
gine. Google and Open-i both query different image
datasets from what we were able to integrate (results
discussed in Section 4.2).
Hybrid (Text and Image) Search Evaluation There
are no public search engines that support text and im-
age radiology queries. IRIS hybrid search was eval-
uated by a survey with four computing experts with
experience in medical imaging retrieval (but without
medical training). We asked the users to indepen-
dently evaluate our results from 0 (“not relevant”) to 2
(“most relevant”) and provided them with evaluation
criteria (using the same coding standards) for text,
image, and hybrid search results. For image search
ranking, our evaluators scored results based on visual
similarity of the images (which was inherently more
subjective than text evaluation).
4 EXPERIMENTAL EVALUATION
4.1 Text Search Query Results
We performed three different types of evaluation: 1)
We compared the merits of different IRIS search fea-
tures in Section 4.1.1. 2) We compared IRIS with
MIRC native search and Google search of the MIRC
site in Section 4.1.2 (since our data comes from MIRC
repository) and 3) We evaluated several other search
engines each using their own datasets in Section 4.1.3.
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
379
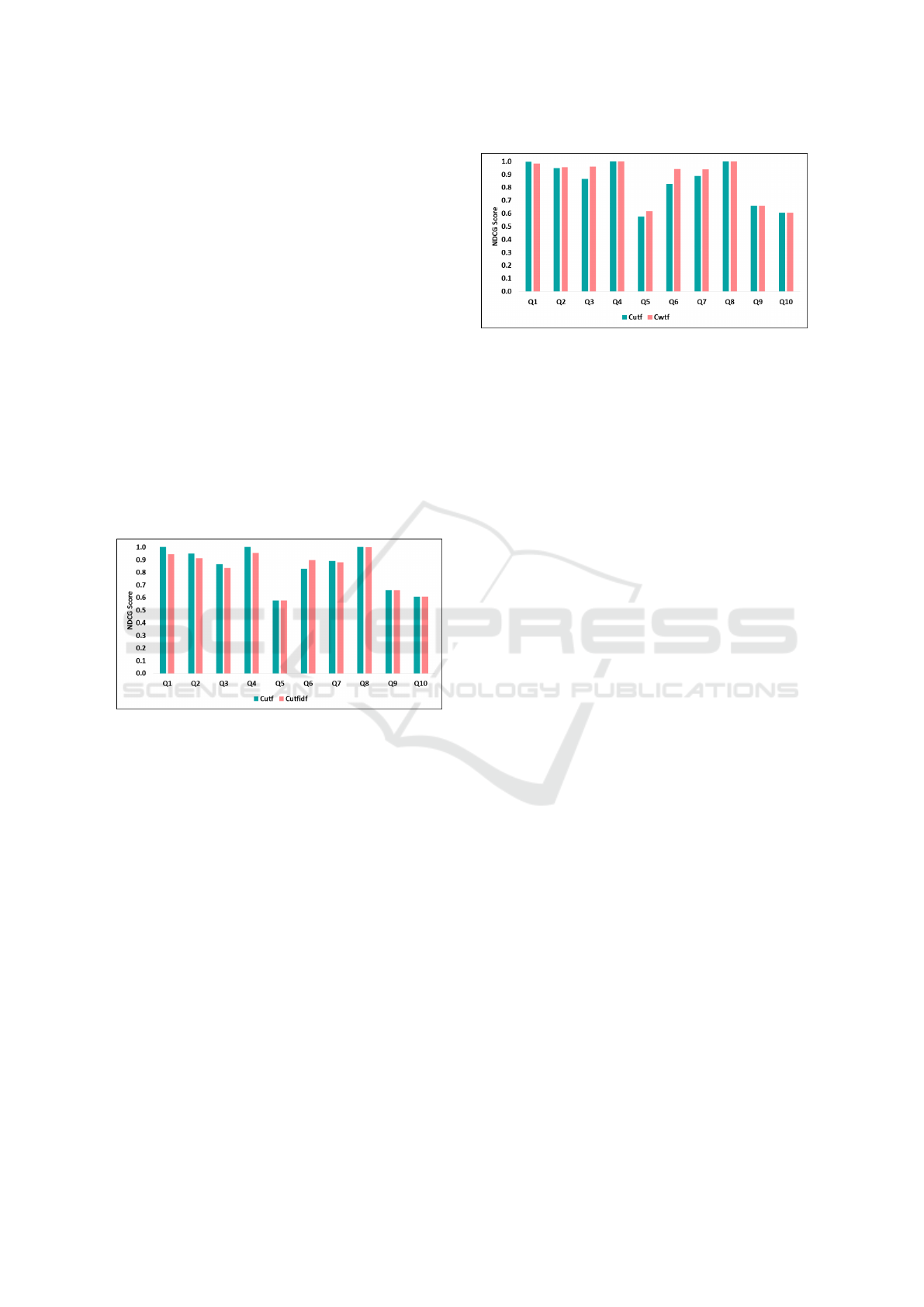
4.1.1 An Evaluation of IRIS Text Search
Our first comparison considers the difference be-
tween unweighted Term Frequency (C
ut f
) and un-
weighted Term Frequency Inverse Document Fre-
quency (C
ut f id f
) discussed in Sections 3.2.1 and 3.2.2.
Our goal is to determine whether adding TF-IDF im-
proves the search results, as TF-IDF is commonly
used in information retrieval algorithms. To compare
TF and TF-IDF we evaluated the NDCG score (on
a 0.0-1.0 scale) for top 10 query results. Figure 5
compares the quality of C
ut f
and C
ut f id f
results. The
queries exhibit minimal variation. The average score
of C
ut f
is 0.84 (standard deviation/SD 0.173) and
C
ut f id f
is 0.82 (SD 0.173) – therefore, using C
ut f id f
is
not providing any benefit in ranking quality. We be-
lieve that C
ut f id f
does not offer any advantage because
domain-specific terms in radiology data do not benefit
from C
ut f id f
normalization – the medical queries for
a specific condition do not need to be normalized by
how common a word is in the corpus.
Figure 5: IRIS text-based search evaluation (NDCG based)
with C
ut f
and C
ut f id f
category.
Our next step considers the advantage of incorpo-
rating term weights (based on data category and on-
tologies) into our search algorithm. Figure 6 shows
the evaluation comparing NDCG scores with un-
weighted term frequency vs weighted term frequency.
The average score of C
ut f
is 0.84 (SD 0.173) and
C
wt f
is 0.87 (SD 0.173). Several query scores are im-
proved, while others remained about the same. We
note that four of the queries (Q1, Q2, Q4, and Q8)
do not have much room for improvement in C
ut f
as
NDCG score cannot exceed 1.
We also compared the performance of unweighted
co-occurrence and weighted TF-IDF. Due to space
constraints we do not present that evaluation. Sim-
ilarly to the comparison between C
ut f
and C
ut f id f
,
we concluded that unweighted co-occurrence func-
tion does not improve ranking quality of IRIS results.
Finally, we compared search relevance scores
from weighted term frequency C
wt f
and weighted
term frequency based co-occurrence C
wc
. The result-
Figure 6: IRIS text-based search evaluation (NDCG based)
with C
ut f
and C
wt f
category.
ing NDCG scores are shown in Figure 7. The average
score of C
wt f
is 0.87 (SD 0.173) and the average score
for C
wc
is 0.96 (SD 0.03). We note that several of the
queries using weighted term frequency have scores
approaching 1.0 and thus a limited scope for improve-
ment. The queries that have relatively poor results
(Q5, Q9, and Q10) have improved significantly, while
queries with near-1 scores remain about the same.
For example, our corpus has many cases involving
explicit mention of cardiomegaly (Q1) in the Diagno-
sis category. In that case, weighted term frequency
can find good matches. We believe that weighted term
frequency and co-occurrence approaches to be com-
plementary – when we have many matching cases,
weighted term frequency ranks results and when we
have few matching cases, co-occurrence finds addi-
tional results based on terms co-occurring with search
terms. For query #2 and #4 (“ACL Tear” and “Pseu-
docoxalgia”) the co-occurrence algorithm benefits
less compared to weighted term frequency because
co-occurring terms with query term return teaching
files where query term does not belong to diagnosis
category (when a query term belongs to diagnosis cat-
egory, it gains more weight). For example, for “ACL
Tear” co-occurring terms are “tear” and “PCL-tear”.
Though our results fetch documents related to “ACL
tear”, those offer a lower relevance rank score com-
pared to weighted term frequency algorithm. How-
ever, the co-occurrence algorithm performs far better
for query (Q5, Q9, and Q10) that returns fewer re-
sults based on the weighted term frequency ranking.
Our average scores of C
wt f
and C
wc
show relative im-
provement in ranking score (based on all 10 queries)
compared to other categories.
4.1.2 Comparison of IRIS and MIRC Search
To compare IRIS relevance rank algorithm with
MIRC, we chose 5 queries (“cardiomegaly”, “ap-
pendicitis”, “tracheal dilation”, “angiosarcoma”, and
“acl tear”). For this experiment, we replaced “no car-
diomegaly” by “appendicitis” because other search
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
380
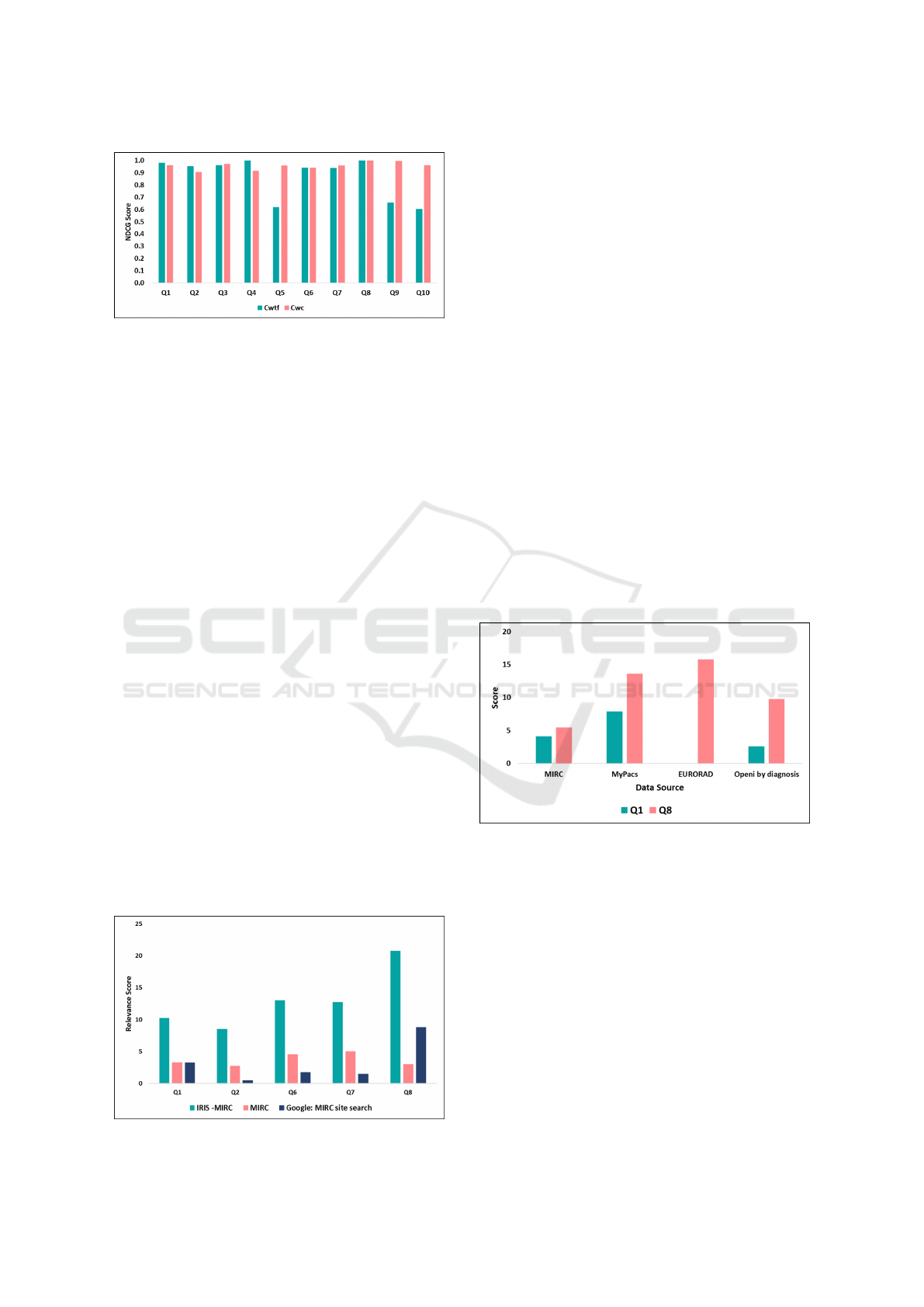
Figure 7: IRIS text-based search evaluation (NDCG based)
with C
wt f
and C
wc
category.
engines do not recognize negation. We considered
top four teaching file results from IRIS, MIRC, and
Google site search. We calculated relevance score
by scoring top four teaching files from each engine,
using our weighted ranking algorithm (discussed in
Section 3.2.1). Figure 8 shows the overall analysis of
results from these search engines. Average score for
each search engine shows that IRIS ranking algorithm
performs better than the other two engines. In that
search, integration of ontologies plays a vital role. For
example, our search matches a teaching file “Ebstein
Anomaly” wherehttps://www.overleaf.com/project/
5cd4382f92a3ba5b77466ce4 cardiomegaly does not
appear in any of the categories; only the synonyms
such as “cardiac enlargement” appear in findings,
“enlarged heart” in differential diagnosis, and
“cardiac enlargement” in the discussion category.
For the “cardiomegaly” search nine teaching cases
tied for the sixth position: “Ventricular septal de-
fect (VSD)”, “Atrioventricular (AV) canal”, “Tricus-
pid atresia” (and 6 others). We used the co-occurrence
algorithm to break the tie and re-rank rest of the re-
sults using a weighted co-occurrence of search terms.
Following top match co-occurrence algorithm ranked
these results as #6. “Ventricular septal defect (VSD)”,
#7. “Tricuspid atresia”, #8.“Atrioventricular (AV)
canal” (and 6 others with lower relevance). We man-
ually verified the accuracy of this ranking based on
presence of co-occurring terms.
Figure 8: IRIS relevance comparison with MIRC.
4.1.3 Ranking Evaluation of Other Medical
Search Engines
We also considered how other public medical radi-
ology teaching file search engines rank their search
results. We used the same query set and performed
a search using MIRC, MyPacs, EURORAD, and
Open-i search engines. We discuss only two out of
five queries (“cardiomegaly” and “appedicitus”) in
detail and report scores for the top 10 search results.
Figure 9 shows a comparative analysis of ranked re-
sults from these four engines using the relevance
scores based on our metric described in Section 3.2.1.
Open-i can rank search results based on different cat-
egories (e.g., based on diagnosis or based on teaching
file date) – we used a diagnosis based search in Open-
i. MIRC ranks results based on the date of modifi-
cation with no other option available. Our analysis
shows that none of the search engines return most rel-
evant results first. Interestingly, top four to five results
are less relevant than the subsequent five results. For
example for “cardiomegaly” MyPacs fourth result is
more relevant than the top three results. EURORAD
does not retrieve any results for “cardiomegaly”; the
results for “appendicits” are also not ranked based on
the relevance of the search term.
Figure 9: Rank retrieval score results from other medical
search engines.
4.2 Image Search Analysis
To evaluate image search, we selected five images
(four images drawn from our database and one image
found using a Google search) associated with our five
chosen text queries. For “no cardiomegaly” query we
used an image from a “normal heart” Google search
(cardiomegaly is a an abnormally enlarged heart);
when we searched for “no cardiomegaly” Google re-
turned “cardiomegaly” images. We chose the other 4
images from our database from one of the top search
results returned by the text based search.
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
381
4.2.1 IRIS-CBIR Evaluation
In this section, we discuss a search for one image out
of the five we evaluate (obtained from a teaching file
with a “cardiomegaly” diagnosis). IRIS retrieved a
total of 13 results based on distance threshold (see
Section 3.3) from teaching files with similar images
like the “cardiomegaly” image. Out of these results
only 3 teaching files were not related to heart diseases;
the remaining 10 teaching cases were about heart dis-
ease diagnosis. We observed that 3 non-heart dis-
ease teaching files had visually similar images as the
search query image (as a teaching file can include im-
ages of different body parts). For example, one of the
returned teaching files contained the diagnosis “Com-
plete duplicated right renal collecting system. Upper
moiety ureter with ectopic ureterocele, Grade V reflux
into lower moiety collecting system.” The images of
the kidney matched the heart search because scanned
images also showed patient’s heart.
4.2.2 Comparison with Other Search Engines
We evaluated all five image queries with the help of
a panel of experts; each reviewer rated the results on
the scale of 0 (not similar) to 4 (very similar).
4.3 Hybrid Search Evaluation
For the hybrid search evaluation we used 5 queries,
although we only discuss one query (based on “car-
diomegaly”) in detail. We applied a threshold of 0.12
to image similarity measure; IRIS text results are
combined with image-based results (as described in
Section 3.4).
We performed hybrid search using “car-
diomegaly” text query and a “cardiomegaly” related
image – the same image that we previously used in
Section 4.2. For “cardiomegaly” query hybrid search
returns 50 cases where text includes “cardiomegaly”
and images are similar to “cardiomegaly” image and
results ranked on the basis of search query relevance.
Using hybrid search, IRIS augmented the text-based
results with image-based results and re-ranked
teaching files based on the latter. Evaluation of the
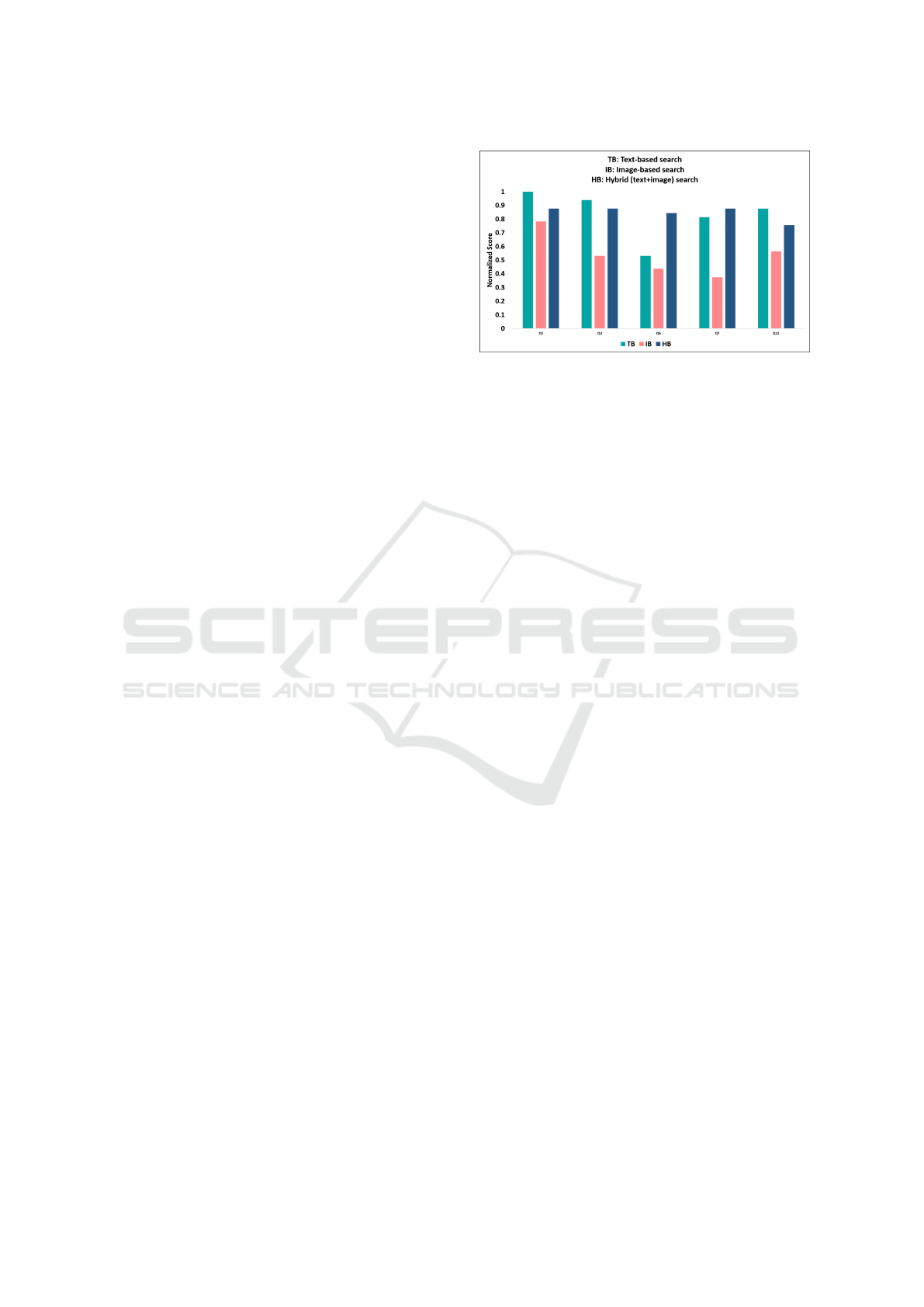
hybrid search using 5 queries is shown in Figure 10.
We defined a relevance criteria and asked evalua-
tors to rank results on the 0-2 scale. We used 5 text
queries and 5 images as input queries, summarized in
Figure 10 (scaled down to 0-1 range for consistency
with other graphs). IRIS text-based and hybrid search
results scored an average score of 0.83 (out of 1) Im-
age search results scored only about 0.53, which fur-
ther confirms our approach of using the image search
as an enhancement to the text search (rather than a
Figure 10: IRIS query evaluation: averaged text, image, and
hybrid results rating with 4 evaluators.
standalone search). Hybrid search produced an aver-
age score of 0.84. IRIS retrieves better results com-
pared to the preliminary results from our earlier work.
We performed a statistical significance test (paired
t-test) for text-based vs image-based search, text-
based vs hybrid search, and image-based vs hybrid
search. The improvement difference was statistically
significant with text-based vs image-based search and
hybrid search vs image-based search. However, hy-
brid search was not a statistically significant improve-
ment over text-based search. One of the reason we
were unable to demonstrate a statistically significant
improvement is a small dataset in this evaluation. As
shown in Figure 10, one of the queries (Q6) was sig-
nificantly improved by the introduction of the hybrid
search, while other four searches hybrid results were
comparable in quality to the original text search.
5 CONCLUSIONS
The ranking approach presented in this paper is sig-
nificant because it enables IRIS to present the user
with the most relevant reference cases first. Through
incorporating term frequency, adding extra weight to
ontology terms, and considering co-occurrence of the
terms, we showed that relevance of teaching file re-
trieval can be improved. In our future work, we plan
to consider the proximity of the terms when calculat-
ing the co-occurrence matrix as well as expand the use
of RadLex and SNOMED CT ontologies from term
synonyms to concepts and categories information and
integrate additional ontologies and data sources.
IRIS will enable radiologists to perform text based
search, image based search as well as hybrid (im-
age and text) searches over integrated datasets. Ul-
timately, IRIS will allow radiologists to make faster,
more confident and accurate diagnoses by removing
the innate error caused by the limits of human mem-
ory. Based on extensive discussions with experienced
KDIR 2019 - 11th International Conference on Knowledge Discovery and Information Retrieval
382

radiologists, IRIS will be a great improvement of
existing search engines – currently radiologists use
in-house teaching file search engines with a limited
search capability.
REFERENCES
Akg
¨
ul, C. B., Rubin, D. L., Napel, S., Beaulieu, C. F.,
Greenspan, H., and Acar, B. (2011). Content-based
image retrieval in radiology: current status and future
directions. Journal of Digital Imaging, 24(2):208–
222.
Channin, D. S., Mongkolwat, P., Kleper, V., Sepukar, K.,
and Rubin, D. L. (2010). The cabig
TM
annotation
and image markup project. Journal of digital imag-
ing, 23(2):217–225.
Dashevsky, B., Gorovoy, M., Weadock, W. J., and Juluru,
K. (2015). Radiology teaching files: an assessment
of their role and desired features based on a national
survey. Journal of digital imaging, 28(4):389–398.
Deshpande, P., Rasin, A., Brown, E., Furst, J., Raicu, D.,
Montner, S., and Armato III, S. (2017). An integrated
database and smart search tool for medical knowledge
extraction from radiology teaching files. In Medical
Informatics and Healthcare, pages 10–18.
Deshpande, P., Rasin, A., Brown, E., Furst, J., Raicu, D. S.,
Montner, S. M., and Armato, S. G. (2018a). Big
data integration case study for radiology data sources.
In 2018 IEEE Life Sciences Conference (LSC), pages
195–198. IEEE.
Deshpande, P., Rasin, A., Brown, E. T., Furst, J., Montner,
S. M., Armato III, S. G., and Raicu, D. S. (2018b).
Augmenting medical decision making with text-based
search of teaching file repositories and medical on-
tologies: Text-based search of radiology teaching
files. International Journal of Knowledge Discovery
in Bioinformatics (IJKDB), 8(2):18–43.
Deshpande, P., Rasin, A., Furst, J., Raicu, D., and Antani,
S. (2019a). Diis: A biomedical data access framework
for aiding data driven research supporting fair princi-
ples. Data, 4(2):54.
Deshpande, P., Rasin, A., Jun, S., Sungmin, K., Brown, E.,
Furst, J., Raicu, D. S., Montner, S. M., and Armato,
S. G. (2019b). Ontology-based radiology teaching
files summarization, coverage, and integration. Jour-
nal of digital imaging, page yet to appear.
Do, B. H., Wu, A., Biswal, S., Kamaya, A., and Rubin,
D. L. (2010). Informatics in radiology: Radtf: A se-
mantic search–enabled, natural language processor–
generated radiology teaching file 1. Radiographics,
30(7):2039–2048.
Dos-Santos, M. and Fujino, A. (2012). Interactive ra-
diology teaching file system: the development of
a mirc-compliant and user-centered e-learning re-
source. In Engineering in Medicine and Biology So-
ciety (EMBC), 2012 Annual International Conference
of the IEEE, pages 5871–5874. IEEE.
Gutmark, R., Halsted, M. J., Perry, L., and Gold, G. (2007).
Use of computer databases to reduce radiograph read-
ing errors. Journal of the American College of Radi-
ology, 4(1):65–68.
Hwang, K. H., Lee, H., Koh, G., Willrett, D., and Rubin,
D. L. (2016). Building and querying rdf/owl database
of semantically annotated nuclear medicine images.
Journal of Digital Imaging, pages 1–7.
Kamauu, A. W. C., DuVall, S. L., Robison, R. J., Liimatta,
A. P., Richard H. Wiggins, I., and Avrin, D. E. (2006).
Vendor-neutral case input into a server-based digital
teaching file system. RadioGraphics, 26(6):1877–
1885. PMID: 17102058.
Kansagra, A. P., John-Paul, J. Y., Chatterjee, A. R.,
Lenchik, L., Chow, D. S., Prater, A. B., Yeh, J., Doshi,
A. M., Hawkins, C. M., Heilbrun, M. E., et al. (2016).
Big data and the future of radiology informatics. Aca-
demic radiology, 23(1):30–42.
Korenblum, D., Rubin, D., Napel, S., Rodriguez, C., and
Beaulieu, C. (2011). Managing biomedical image
metadata for search and retrieval of similar images.
Journal of digital imaging, 24(4):739–748.
Li, Y., Liang, X., Hu, Z., and Xing, E. P. (2018). Hybrid
retrieval-generation reinforced agent for medical im-
age report generation. In Advances in Neural Infor-
mation Processing Systems, pages 1537–1547.
Ling, Z. J., Tran, Q. T., Fan, J., Koh, G. C., Nguyen, T., Tan,
C. S., Yip, J. W., and Zhang, M. (2014). Gemini: an
integrative healthcare analytics system. Proceedings
of the VLDB Endowment, 7(13):1766–1771.
M
¨
uller, H., M
¨
uller, W., Squire, D. M., Marchand-Maillet,
S., and Pun, T. (2001). Performance evaluation in
content-based image retrieval: overview and propos-
als. Pattern recognition letters, 22(5):593–601.
Pinho, E., Godinho, T., Valente, F., and Costa, C. (2017). A
multimodal search engine for medical imaging stud-
ies. Journal of digital imaging, 30(1):39–48.
Russell-Rose, T. and Chamberlain, J. (2017). Expert
search strategies: The information retrieval practices
of healthcare information professionals. JMIR, 5(4).
Simpson, M. S., Demner-Fushman, D., Antani, S. K., and
Thoma, G. R. (2014). Multimodal biomedical image
indexing and retrieval using descriptive text and global
feature mapping. Information retrieval, 17(3):229–
264.
Talanow, R. (2009). Radiology teacher: a free, internet-
based radiology teaching file server. Journal of the
American College of Radiology, 6(12):871–875.
Thies, C., G
¨
uld, M. O., Fischer, B., and Lehmann, T. M.
(2004). Content-based queries on the casimage
database within the irma framework. In Workshop of
the Cross-Language Evaluation Forum for European
Languages, pages 781–792. Springer.
Tsikrika, T., de Herrera, A. G. S., and M
¨
uller, H. (2011).
Assessing the scholarly impact of imageclef. In
Forner, P., Gonzalo, J., Kek
¨
al
¨
ainen, J., Lalmas, M.,
and de Rijke, M., editors, Multilingual and Multi-
modal Information Access Evaluation, pages 95–106.
Springer Berlin Heidelberg.
Weinberger, E., Jakobovits, R., and Halsted, M. (2002).
Mypacs. net: a web-based teaching file authoring tool.
American Journal of Roentgenology, 179(3):579–582.
Multimodal Ranked Search over Integrated Repository of Radiology Data Sources
383
